 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sp_144720_excs.t2 | ||||||||
| Common Name | SOVF_144720 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Chenopodiaceae; Chenopodioideae; Anserineae; Spinacia
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 719aa MW: 79370 Da PI: 5.0217 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 40.6 | 5.7e-13 | 666 | 715 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHT...TTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmg...kgRtlkqcksrwqkyl 48
r++W+ eE++ l avk+lG + Wk+I ++ g +gR+ ++k++w++ l
Sp_144720_excs.t2 666 RKKWSVEEEDTLRVAVKELGRN-WKLILQCHGdafEGRSNVDLKDKWRNML 715
789*****************66.*************************975 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 13.596 | 663 | 719 | IPR017930 | Myb domain |
| SMART | SM00717 | 5.5E-10 | 665 | 717 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.6E-10 | 666 | 715 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.4E-12 | 666 | 719 | IPR009057 | Homeodomain-like |
| CDD | cd11660 | 8.80E-13 | 667 | 716 | No hit | No description |
| SuperFamily | SSF46689 | 3.26E-12 | 668 | 717 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009901 | Biological Process | anther dehiscence | ||||
| GO:0010152 | Biological Process | pollen maturation | ||||
| GO:0043067 | Biological Process | regulation of programmed cell death | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 719 aa Download sequence Send to blast |
MDEKVAKWII EFLLRKPIDE KIINGVLSCL PLSNNDKRLK KTILLRKIES AISDGSVSEK 60 LLELLEMVDE LNHGAGKTVP DSMKRAYCAV AVDCTVRFLE ENVEENGKYF EAVERVWRDR 120 VCKVESLVSE ELNRWLEDIE AAVWDSSVCE KILMRNTRNE ALRAVKRYLK EAWESMGPSF 180 LELVALKVGN DVGELCRSEG GGGGDGGGGG GSGSSGGGGS GGVVGSDNPC RTLIDPQGLV 240 ESQAFNRRKH VAVKTRRPRG IKIFDAVEAA ETSMNTANDI QTREMDKVQE TLQPSSTQQH 300 GAANDSHPST SRTAHNLSCG KNNDAPIYKM DKVQEALQPS STQLHGAVND SHPSTSRTAR 360 NLSCGKDNDA PIFKMDKVQE ALESNSFHLH ADVSDSHLDP LEAVHNMSVT KVTDNMSNLN 420 AIKKQTTHET PEVRRVQEAL NFSARELHEA VTDPLPEALE IAKTLSSAVG ATLNHGTGHV 480 TGVENQNTSN DDVLENQTTS NDDVLENQNT GNDGVFENHN TGSDTVFENH NTGNDAVLEN 540 QNTGNDAVLE NRNTGNDDVL ENQIQPPVIH ATEKNTENVE EEMPELAGDA QGNNETDDPK 600 KQHRSKRSLM EANGTSHTFE WDDSIDNVNR GSELHLPPSP KRSKVTPLEQ YEDKPKPKPQ 660 PLIKGRKKWS VEEEDTLRVA VKELGRNWKL ILQCHGDAFE GRSNVDLKDK WRNMLRSQL |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00052 | PBM | Transfer from AT1G15720 | Download |
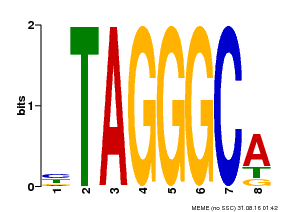
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021857414.1 | 0.0 | protein CLEC16A homolog isoform X2 | ||||
| TrEMBL | A0A0K9QSX4 | 0.0 | A0A0K9QSX4_SPIOL; Uncharacterized protein | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G15720.1 | 2e-17 | TRF-like 5 | ||||



