 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sp_157270_hzna.t1 | ||||||||
| Common Name | SOVF_157270 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Chenopodiaceae; Chenopodioideae; Anserineae; Spinacia
|
||||||||
| Family | FAR1 | ||||||||
| Protein Properties | Length: 848aa MW: 97101.7 Da PI: 6.9851 | ||||||||
| Description | FAR1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | FAR1 | 82.4 | 7.5e-26 | 88 | 189 | 2 | 91 |
FAR1 2 fYneYAkevGFsvrkskskkskrngeitkrtfvCskegkreeekkk............tekerrtraetrtgCkaklkvkkekdgkwevtkl 81
fY+eYA+++GF++ +++s++sk+++e+++++f Cs++g+++e +k e++ +r+ +t+Cka+++vk++ dg+w+++++
Sp_157270_hzna.t1 88 FYQEYARSMGFNTAIQNSRRSKTSREFIDAKFACSRYGTKREYDKGnrprirqcnkqePESATGRRSCGKTNCKASMHVKRRPDGRWVIHSF 179
9**************************************999998889999999775555555589999*********************** PP
FAR1 82 eleHnHelap 91
+eHnHel p
Sp_157270_hzna.t1 180 IKEHNHELLP 189
*******975 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03101 | 1.3E-23 | 88 | 189 | IPR004330 | FAR1 DNA binding domain |
| Pfam | PF10551 | 9.6E-29 | 287 | 379 | IPR018289 | MULE transposase domain |
| PROSITE profile | PS50966 | 8.951 | 568 | 604 | IPR007527 | Zinc finger, SWIM-type |
| SMART | SM00575 | 7.9E-6 | 579 | 606 | IPR006564 | Zinc finger, PMZ-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009585 | Biological Process | red, far-red light phototransduction | ||||
| GO:0010218 | Biological Process | response to far red light | ||||
| GO:0042753 | Biological Process | positive regulation of circadian rhythm | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 848 aa Download sequence Send to blast |
MDIDLRLPSG EEDLQDEDQT TDSNMLDRYA KLDSSGDAET TLTVAEVHAE EELALGSTTS 60 DLAEYKEDVN LEPLHGMEFV SHQEAYAFYQ EYARSMGFNT AIQNSRRSKT SREFIDAKFA 120 CSRYGTKREY DKGNRPRIRQ CNKQEPESAT GRRSCGKTNC KASMHVKRRP DGRWVIHSFI 180 KEHNHELLPA QAVSEQTRKM YAAMARNYAE YKNVVGLKND SKIPFDKNRN LLLDAGDAKL 240 LLEFFTQMQT VNSNFFYAVD LSDDLRLKSL LWIDAKSRHD YVNFNDVVSF DTAYIRNKYK 300 MPLALLVGVN QHYQFMLLGC AIVSDESTAT YSWILQTWLK AMGGQAPKVI ITDQDDAIKS 360 AVSEVFPITR HCFHLWHILG KVSENLGHII KKHENFMAKF DKCIYRSWGD EEFEKRWQKL 420 VVRFELNKDD EWIQSLYEER KLWVPAFMKD CFFAGMSAIQ RSDSVNSYFD KYVHKKTAVP 480 DFLKQYDTIT QDRYEEEARA DSDTWNKQPA LKSPSPFEKH MSTLYTHAMF RKFQVEVLGA 540 VACHPKRERQ EDLITTFKVQ DFEKNQDFLV TWNEMKSEVS CICHLFEYKG FLCRHTMIVL 600 QICGLSMIPS QYILKRWTKD AKERHLIGDG TEQVLSRVQR YNDLCNRAIK LGEEGSLSQE 660 SYNLALRALD EAFTNCIGVN NSCKTLVEVG TSATHGLLSI EDGSPSRSFS KTAKKKNPTK 720 KRKMNTETEA MAIGSQDGLQ QMDKLGSRPV TTLDGFYGTQ QGVQGMVQLN LMAPTRDNYY 780 GNPPAIQGLG QLNSIAPSHE GYFGQQTMHG LGQMDFFRGP TGFSYGIREE ANVNVRPAQL 840 HEDGARHS |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
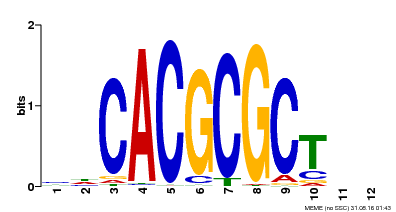
| UniProt | Transcription activator that recognizes and binds to the DNA consensus sequence 5'-CACGCGC-3'. Activates the expression of FHY1 and FHL involved in light responses. When associated with PHYA, protects it from being recognized and degraded by the COP1/SPA complex. Positive regulator of chlorophyll biosynthesis via the activation of HEMB1 gene expression. {ECO:0000269|PubMed:11889039, ECO:0000269|PubMed:12753585, ECO:0000269|PubMed:17012604, ECO:0000269|PubMed:18033885, ECO:0000269|PubMed:18715961, ECO:0000269|PubMed:22634759}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00078 | ChIP-seq | Transfer from AT3G22170 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated after exposure to far-red light. Subject to a negative feedback regulation by PHYA signaling. Up-regulated by white light. {ECO:0000269|PubMed:11889039, ECO:0000269|PubMed:18033885, ECO:0000269|PubMed:22634759}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021843597.1 | 0.0 | protein FAR-RED ELONGATED HYPOCOTYL 3 | ||||
| Refseq | XP_021843598.1 | 0.0 | protein FAR-RED ELONGATED HYPOCOTYL 3 | ||||
| Refseq | XP_021843599.1 | 0.0 | protein FAR-RED ELONGATED HYPOCOTYL 3 | ||||
| Swissprot | Q9LIE5 | 0.0 | FHY3_ARATH; Protein FAR-RED ELONGATED HYPOCOTYL 3 | ||||
| TrEMBL | A0A0K9QP72 | 0.0 | A0A0K9QP72_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010680156.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G22170.2 | 0.0 | far-red elongated hypocotyls 3 | ||||



