 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sp_158140_zshu.t1 | ||||||||
| Common Name | SOVF_158140 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Chenopodiaceae; Chenopodioideae; Anserineae; Spinacia
|
||||||||
| Family | C3H | ||||||||
| Protein Properties | Length: 348aa MW: 38920.7 Da PI: 7.3273 | ||||||||
| Description | C3H family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-CCCH | 43.8 | 4.3e-14 | 271 | 296 | 1 | 26 |
--S---SGGGGTS--TTTTT-SS-SS CS
zf-CCCH 1 yktelCrffartGtCkyGdrCkFaHg 26
+ktelC+ +++t tC+yGd C+FaHg
Sp_158140_zshu.t1 271 FKTELCNKWQETKTCPYGDHCQFAHG 296
79***********************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1000.10 | 1.5E-18 | 265 | 303 | IPR000571 | Zinc finger, CCCH-type |
| SuperFamily | SSF90229 | 5.49E-11 | 267 | 302 | IPR000571 | Zinc finger, CCCH-type |
| SMART | SM00356 | 6.0E-8 | 270 | 297 | IPR000571 | Zinc finger, CCCH-type |
| PROSITE profile | PS50103 | 15.478 | 270 | 298 | IPR000571 | Zinc finger, CCCH-type |
| Pfam | PF00642 | 3.3E-11 | 271 | 296 | IPR000571 | Zinc finger, CCCH-type |
| SuperFamily | SSF90229 | 3.4E-8 | 304 | 337 | IPR000571 | Zinc finger, CCCH-type |
| Gene3D | G3DSA:4.10.1000.10 | 7.2E-12 | 304 | 334 | IPR000571 | Zinc finger, CCCH-type |
| SMART | SM00356 | 1.3E-4 | 308 | 335 | IPR000571 | Zinc finger, CCCH-type |
| PROSITE profile | PS50103 | 12.878 | 308 | 336 | IPR000571 | Zinc finger, CCCH-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 348 aa Download sequence Send to blast |
MQQDTPASTM MMNQMSSAAD IFSPPPYSSI FFHSATSPTT FKASPCTSEG QSTINAASFA 60 TTAAKHLSRL MSDHKELLSR QSLCLAQLRK TFKEAETLRQ ENTNLRLANV ELSNQVNLLI 120 QATLQSQYQA VPFSDYPEQY GSSSLSAAAE KFSVGVDGCG FEKERECEDW DEGFYSKNQN 180 QSSVIEKNLV EDHKEENDRV SLPKSISVRS NGFLKISQIA AAPTRKVSAP TQSVTRIGSP 240 NPVSGPQKVY VRGGSKKEGP IELEVYNQGM FKTELCNKWQ ETKTCPYGDH CQFAHGIDEL 300 RPVIRHPRYK TEVCRMVLSG DSCPYGHRCH FRHALTDEEK LMHPHMHN |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1rgo_A | 3e-20 | 271 | 333 | 4 | 66 | Butyrate response factor 2 |
| Search in ModeBase | ||||||
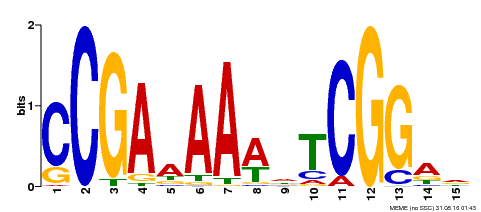
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00383 | ampDAP | Transfer from AT1G68200 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021844986.1 | 0.0 | zinc finger CCCH domain-containing protein 15 | ||||
| TrEMBL | A0A0K9QNV4 | 0.0 | A0A0K9QNV4_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010681714.1 | 1e-156 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G68200.1 | 4e-80 | C3H family protein | ||||



