 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sp_160140_egdw.t2 | ||||||||
| Common Name | SOVF_160140 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Chenopodiaceae; Chenopodioideae; Anserineae; Spinacia
|
||||||||
| Family | C3H | ||||||||
| Protein Properties | Length: 622aa MW: 67940.9 Da PI: 6.1312 | ||||||||
| Description | C3H family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-CCCH | 21.3 | 4.6e-07 | 264 | 284 | 5 | 26 |
--SGGGGTS--TTTTT-SS-SS CS
zf-CCCH 5 lCrffartGtCkyGdrCkFaHg 26
+C+ f++ G C+ Gd C +aHg
Sp_160140_egdw.t2 264 PCPEFRK-GSCPKGDVCEYAHG 284
8******.*************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF48403 | 2.02E-14 | 13 | 139 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 3.2E-16 | 15 | 177 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 2.5E-9 | 58 | 130 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 0.0081 | 62 | 92 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50297 | 16.175 | 62 | 137 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 1.09E-12 | 64 | 137 | No hit | No description |
| SMART | SM00248 | 0.007 | 97 | 129 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 11.728 | 97 | 132 | IPR002110 | Ankyrin repeat |
| SMART | SM00356 | 9.4E-5 | 259 | 285 | IPR000571 | Zinc finger, CCCH-type |
| Gene3D | G3DSA:4.10.1000.10 | 5.1E-4 | 263 | 286 | IPR000571 | Zinc finger, CCCH-type |
| PROSITE profile | PS50103 | 12.389 | 264 | 286 | IPR000571 | Zinc finger, CCCH-type |
| SMART | SM00356 | 56 | 294 | 317 | IPR000571 | Zinc finger, CCCH-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 622 aa Download sequence Send to blast |
MESEDYKKDT IRDHQTSLPL LELSATNDLS SFKDAVEKEG YDLDEASLWY GRRVGSKKMG 60 FEERTPLMVA AMFGSKDILN YILQTSLVDV NRASGSDGAT ALHCAAAGGS LYSHEVIKLL 120 VESSANVDQV DVHGNRPCDV IAPCFSFSSK LKRRTLDILL NKGDEDEDSS DQTEEGIQDL 180 HLEGFSPKSS SDGSGSERKE YPIDPSLPDI KNGIYGTDEF RMYTFKVKPC SRAYSHDWTE 240 CPFVHPGENA RRRDPRKYHY SCVPCPEFRK GSCPKGDVCE YAHGIFECWL HPAQYRTRLC 300 KDEISCARRV CFFAHRPEEL RPLYASTGSA VPSPRSYSSS GPSFDMGSLS PLALGSPSML 360 MPSTSTPPMT PTGASSPIGG AAAWQNHLNI TPPPALQLPG SRLKAALSSR DLGMETGLRR 420 QQLLDELATG VLGRLGGLSL SSPTGVQIRQ NMNQQLLSSY SSGMPSSPVR GSCSSFGMEP 480 GGGSPISNPR LAAFANRSQS FVDRNAVNRL SGGLASPFSH ISDWGSPDGK VDWGVQGDEL 540 NKFRKSASFA YRSSNGNNNS MRGPNGEPDV SWVHSLVKDA PPVNSSSTQH VFEEQYSLNS 600 GMLERIPPWV EQLYMEQEPM AA |
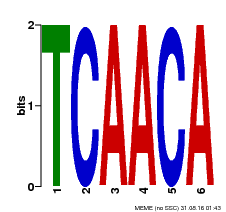
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00566 | DAP | Transfer from AT5G58620 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021852509.1 | 0.0 | zinc finger CCCH domain-containing protein 66-like isoform X1 | ||||
| Swissprot | Q9LUZ4 | 0.0 | C3H66_ARATH; Zinc finger CCCH domain-containing protein 66 | ||||
| TrEMBL | A0A0K9QQB9 | 0.0 | A0A0K9QQB9_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010685536.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G58620.1 | 1e-178 | C3H family protein | ||||



