 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sp_172040_pmpz.t1 | ||||||||
| Common Name | SOVF_172040 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Chenopodiaceae; Chenopodioideae; Anserineae; Spinacia
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 394aa MW: 43553.6 Da PI: 4.6596 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 59.5 | 7.9e-19 | 103 | 153 | 2 | 56 |
AP2 2 gykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkklege 56
+y+GVr++ +g+WvAeIr+p + r kr +lg++ ta eAa +++a+++++ge
Sp_172040_pmpz.t1 103 KYRGVRQRT-WGKWVAEIREP---N-RgKRLWLGTYPTALEAALSYDEAARAMYGE 153
7*****999.**********8...3.35*************************996 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00380 | 2.1E-35 | 103 | 166 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 1.9E-13 | 103 | 153 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 6.54E-20 | 103 | 161 | IPR016177 | DNA-binding domain |
| PROSITE profile | PS51032 | 21.733 | 103 | 160 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 2.6E-29 | 103 | 160 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 2.94E-30 | 103 | 161 | No hit | No description |
| PRINTS | PR00367 | 1.3E-8 | 104 | 115 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.3E-8 | 126 | 142 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0010286 | Biological Process | heat acclimation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 394 aa Download sequence Send to blast |
SSYVLGDFRK IVNEVVVSGA VETGFDSNLH RPCLNYSFQR RRKTRRRCDG TDSLADTLAK 60 WKEINEKLCE QKEDGKPVRK APAKGSKKGC MKGKGGPENS LFKYRGVRQR TWGKWVAEIR 120 EPNRGKRLWL GTYPTALEAA LSYDEAARAM YGESARLNLP YYPAPVREDS RDDSASASYT 180 TATTTSASYT TACTTSGNNS DVYVGEQWDG VTGGATVKRE DIEGDSEFDF KNASAGFKVG 240 TPGTPVDKEE KAVVDNGEGD QGGIDINDYL HNLTMDEMFD VDELLGAIDA GPVSPVGNTT 300 TQGHDAYGAF RSQVENNSIQ LERPESWSNH EQNPNYFHLF QNQDDVNLYG SVNNSDQGYN 360 GVNYGFDFLN PDRQEDSSLP VEDHGFLNLG ELGF |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gcc_A | 7e-16 | 104 | 160 | 3 | 60 | ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 1 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 38 | 43 | QRRRKT |
| 2 | 38 | 45 | QRRRKTRR |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00302 | DAP | Transfer from AT2G40340 | Download |
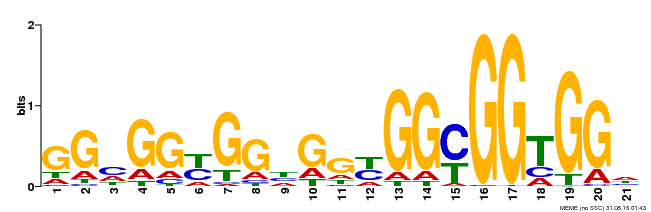
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021855823.1 | 0.0 | dehydration-responsive element-binding protein 2C-like isoform X2 | ||||
| TrEMBL | A0A0K9QLF6 | 0.0 | A0A0K9QLF6_SPIOL; Uncharacterized protein (Fragment) | ||||
| STRING | XP_010692919.1 | 1e-136 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G40340.1 | 2e-38 | ERF family protein | ||||



