 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sp_174400_uesu.t2 | ||||||||
| Common Name | SOVF_174400 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Chenopodiaceae; Chenopodioideae; Anserineae; Spinacia
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 931aa MW: 105947 Da PI: 7.9062 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 164.7 | 1.6e-51 | 29 | 146 | 2 | 118 |
CG-1 2 lke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenp 92
+ke k+rwl+++ei+aiL n+++++++ ++ + p+ g+++L++rkk+r+frkDG++wkkkkdgktv+E+he+LKvg+ e +++yYah+e+np
Sp_174400_uesu.t2 29 MKEaKSRWLRPNEIHAILFNYTNFTIHVKPVSLPRGGTIVLFDRKKLRNFRKDGHNWKKKKDGKTVKEAHEHLKVGTEERIHVYYAHGEDNP 120
45559*************************************************************************************** PP
CG-1 93 tfqrrcywlLeeelekivlvhylevk 118
+f rrcywlL+++le+ivlvhy+e++
Sp_174400_uesu.t2 121 KFVRRCYWLLDKALEHIVLVHYRETQ 146
***********************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 75.689 | 25 | 151 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 1.3E-71 | 28 | 146 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 2.4E-45 | 31 | 144 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 5.74E-12 | 383 | 469 | IPR014756 | Immunoglobulin E-set |
| CDD | cd00204 | 1.84E-15 | 566 | 676 | No hit | No description |
| Gene3D | G3DSA:1.25.40.20 | 4.2E-15 | 566 | 679 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 7.31E-16 | 567 | 679 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 13.39 | 575 | 649 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 2.9E-5 | 617 | 646 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 9.858 | 617 | 649 | IPR002110 | Ankyrin repeat |
| Pfam | PF13637 | 8.7E-5 | 619 | 674 | No hit | No description |
| SuperFamily | SSF52540 | 4.51E-7 | 730 | 831 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| PROSITE profile | PS50096 | 7.272 | 765 | 791 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 20 | 780 | 802 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.492 | 781 | 810 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 2.0E-4 | 803 | 825 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.56 | 804 | 828 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 2.1E-4 | 805 | 825 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 13 | 883 | 905 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.297 | 885 | 913 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 931 aa Download sequence Send to blast |
MESGFNGGLG GYDIHGFRTM EDLDVPNIMK EAKSRWLRPN EIHAILFNYT NFTIHVKPVS 60 LPRGGTIVLF DRKKLRNFRK DGHNWKKKKD GKTVKEAHEH LKVGTEERIH VYYAHGEDNP 120 KFVRRCYWLL DKALEHIVLV HYRETQESQV SPSTPTNSSF RSDLTDYSAS QLLSRGNDCV 180 VNQVYYPSMK ERTEFGDGVS MNNLDMRLHE INTLDWDELL VSNNPTEATL TTREQNPYLQ 240 QDKQPSTFSS ENNGSSLLTG HPHPGMLTSR NSMNPVADTS LAQVTHVGDM FLPLTVVQTH 300 QKEEGRIYGM GAVGAGDLSD KMAKDGLQSQ DSFGKWMNEI IVDSPESVGD PSFESSLETS 360 HGSLMSSGAV NHEGPYPAQI FCITDISPTW AYSTEETKIL VVGFYHQEYR QLAKATVYCV 420 CGDTCVPAEI IQVGVFRCMV SPQSPGSVNF YLSIDCSTPI SQVLTFEFRS PAMTNPVVRQ 480 DKSQWDMFRI QMRLAYLLFT TSKSLDILSS KVSQSALKEG KKFALKYSNT ADSWAYFTKL 540 TESGKITFER AKDNLFELSM KSRLKEWLLE RVVGGSKISE RDAEGQGVLH LCAILDYTWA 600 VYPFSCCGLS LDFRDKFGWT ALHWAAYYGR EKMVAALLSA RAKPNLVTDP TSENPGGCTS 660 ADLAAKQGFE GLAAYLSEKA LVQQFEDMRI AGNAGGSLET QTYETSNANN ITEEELDLKD 720 TLTAYRTAAD AAARIQVAFR EQTLKQRSKI VEFLNPETEA QYIVAAMKIQ HAFRNYELRK 780 QMAAALRIQH RFRTWKLRKD FLNMRRKVIK IQAAFRGFLL RQQYQKFVWS VGVLEKAILR 840 WRLRRKGFRG LKVEIQEPVD DQRQESDTEE DFYRASRKQA EERVEKAVVR VQSMFRSKQA 900 QQEYRRMKLA HTQAQLEFED SVNPNGFMNE M |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 71 | 88 | RKKLRNFRKDGHNWKKKK |
| 2 | 72 | 87 | KKLRNFRKDGHNWKKK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
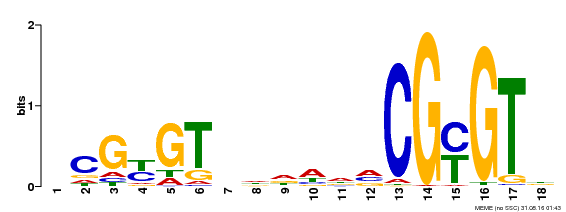
| UniProt | Transcription activator (PubMed:14581622). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in response to cold. Contributes together with CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:28351986, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, wounding, abscisic acid, H(2)O(2) and salicylic acid. {ECO:0000269|PubMed:12218065}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021835012.1 | 0.0 | calmodulin-binding transcription activator 5 isoform X2 | ||||
| Swissprot | O23463 | 0.0 | CMTA5_ARATH; Calmodulin-binding transcription activator 5 | ||||
| TrEMBL | A0A0K9QKS6 | 0.0 | A0A0K9QKS6_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010671665.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 0.0 | calmodulin binding;transcription regulators | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



