 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sp_182220_jxzj.t1 | ||||||||
| Common Name | SOVF_182220 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Chenopodiaceae; Chenopodioideae; Anserineae; Spinacia
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 690aa MW: 78349.7 Da PI: 6.3455 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 150.4 | 1.5e-46 | 82 | 213 | 3 | 137 |
DUF822 3 sgrkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpleeaeaagssasaspessl 94
+gr++++kE+E++k+RER+RRai+++++aGLR++Gn++lp+raD+n+Vl+AL+reAGw+ve+DGttyr + p ++ g +s s+l
Sp_182220_jxzj.t1 82 KGRREREKEKERTKLRERHRRAITSRMLAGLRQYGNFHLPARADMNDVLAALAREAGWMVEPDGTTYRSSPPP----SQLGGLPVRSSDSPL 169
79******************************************************************44333....466677777777777 PP
DUF822 95 q.sslkssalaspvesysaspksssfpspssldsislasaasll 137
+ sslk+++++ ++ + +++ ++ +sp+slds+ +++ ++
Sp_182220_jxzj.t1 170 SaSSLKNCSMKPSLNCQPTVLGIDENLSPASLDSVVVTEMETKN 213
77899******************************998773333 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 1.8E-47 | 82 | 223 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| SuperFamily | SSF51445 | 2.21E-155 | 247 | 684 | IPR017853 | Glycoside hydrolase superfamily |
| Gene3D | G3DSA:3.20.20.80 | 4.9E-167 | 249 | 681 | IPR013781 | Glycoside hydrolase, catalytic domain |
| Pfam | PF01373 | 8.2E-78 | 273 | 638 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.8E-53 | 286 | 300 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.8E-53 | 307 | 325 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.8E-53 | 329 | 350 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.8E-53 | 422 | 444 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.8E-53 | 495 | 514 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.8E-53 | 529 | 545 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.8E-53 | 546 | 557 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.8E-53 | 564 | 587 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.8E-53 | 602 | 624 | IPR001554 | Glycoside hydrolase, family 14 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000272 | Biological Process | polysaccharide catabolic process | ||||
| GO:0048831 | Biological Process | regulation of shoot system development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0016161 | Molecular Function | beta-amylase activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 690 aa Download sequence Send to blast |
MNGAGEDTTP ATFDHHEIHQ NPLNSNLQIP IPGPNPNINN FSHSNSHQYH LISPPQPQPQ 60 TNRRPRGFAV TAGNSSGSGD VKGRREREKE KERTKLRERH RRAITSRMLA GLRQYGNFHL 120 PARADMNDVL AALAREAGWM VEPDGTTYRS SPPPSQLGGL PVRSSDSPLS ASSLKNCSMK 180 PSLNCQPTVL GIDENLSPAS LDSVVVTEME TKNEKNTSTS NIDSPESLEA HQLIQHVQSN 240 LCENDYSGTP HVPVYIKLSS GIINSYCQLV DPEGIRQELK HMKSLNVDGV VVDCWWGIVE 300 GWSPQKYTWL GYRELFMIIR EFQLKVQVVL AFHEYGGSDS GNVFISLPHW VLEIGKDNQD 360 IFFTDREGRR NTECISWGID KERVLRGRTG IEVYFDLMRS FRSEFDDIFA DDLITAVEIG 420 LGASGELKFP SFPERLGWRY PGIGEFQCYD RYLQQNLRKA AKLRGHSFWA RGPDNAGHYN 480 SRPPETGFFC EKGDYESYFG RFFLHWYAQM LIDHADNVLS LASLAFEETQ IIVKIPAVYW 540 WYKTPSHAAE LTAGYYNPTN RDGYSPVFEV FKKHSVAVKY VCPRSRIHCR ENDESFSDPE 600 SLSWQVLSAA WDCGLTIAGQ NADPCFDRGE HIKMIETSKP NNDPDRRHFS FFVYQLPSPF 660 VQRTYCYSEL DNFIKNMHGE NGCHEEPAQV |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wdq_A | 1e-119 | 251 | 689 | 11 | 453 | Beta-amylase |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00543 | DAP | Transfer from AT5G45300 | Download |
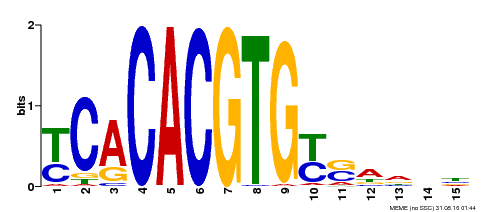
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021858576.1 | 0.0 | beta-amylase 8 | ||||
| Swissprot | Q9FH80 | 0.0 | BAM8_ARATH; Beta-amylase 8 | ||||
| TrEMBL | A0A0K9QGA3 | 0.0 | A0A0K9QGA3_SPIOL; Beta-amylase | ||||
| STRING | XP_010676684.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G45300.1 | 0.0 | beta-amylase 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



