 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sp_185680_rszf.t1 | ||||||||
| Common Name | SOVF_185680 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Chenopodiaceae; Chenopodioideae; Anserineae; Spinacia
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 1377aa MW: 154117 Da PI: 8.3893 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 13.7 | 0.00019 | 1286 | 1308 | 3 | 23 |
ET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 3 Cp..dCgksFsrksnLkrHirtH 23
Cp Cgk F ++ +L++H r+H
Sp_185680_rszf.t1 1286 CPvkGCGKKFFSHKYLVQHRRVH 1308
9999*****************99 PP
| |||||||
| 2 | zf-C2H2 | 12.8 | 0.00035 | 1344 | 1370 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH..T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt..H 23
y+C+ Cg++F+ s++ rH r+ H
Sp_185680_rszf.t1 1344 YVCNepGCGQTFRFVSDFSRHKRKtgH 1370
99********************99666 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00545 | 1.1E-16 | 25 | 66 | IPR003349 | JmjN domain |
| PROSITE profile | PS51183 | 13.91 | 26 | 67 | IPR003349 | JmjN domain |
| Pfam | PF02375 | 7.0E-14 | 27 | 60 | IPR003349 | JmjN domain |
| SMART | SM00558 | 3.6E-49 | 195 | 364 | IPR003347 | JmjC domain |
| PROSITE profile | PS51184 | 33.561 | 198 | 364 | IPR003347 | JmjC domain |
| SuperFamily | SSF51197 | 7.42E-25 | 211 | 378 | No hit | No description |
| Pfam | PF02373 | 1.3E-36 | 228 | 347 | IPR003347 | JmjC domain |
| SuperFamily | SSF57667 | 1.9E-6 | 1282 | 1321 | No hit | No description |
| PROSITE profile | PS50157 | 12.445 | 1284 | 1313 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0045 | 1284 | 1308 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 7.8E-6 | 1285 | 1307 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 1286 | 1308 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 2.2E-9 | 1308 | 1336 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 10.72 | 1314 | 1343 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0023 | 1314 | 1338 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 1316 | 1338 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 1.56E-9 | 1324 | 1367 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 1.5E-9 | 1337 | 1367 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.14 | 1344 | 1370 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 11.365 | 1344 | 1375 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 1346 | 1370 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009741 | Biological Process | response to brassinosteroid | ||||
| GO:0009826 | Biological Process | unidimensional cell growth | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0033169 | Biological Process | histone H3-K9 demethylation | ||||
| GO:0035067 | Biological Process | negative regulation of histone acetylation | ||||
| GO:0040010 | Biological Process | positive regulation of growth rate | ||||
| GO:0045815 | Biological Process | positive regulation of gene expression, epigenetic | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0071558 | Molecular Function | histone demethylase activity (H3-K27 specific) | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1377 aa Download sequence Send to blast |
MGSSSIVTAE HSSPEVFPWL KTLPVAPEYH PTIQEFQDPI AYIFKIEKEA SKFGICKIVP 60 PVPPQSKKSV ISNLNQSLLN SSKNPSNPQP TFTTRQQQVG FCPRKQRPIQ KSVWQSGECY 120 TLQQFEAKAK AFEKNYLKNR AKKAVSPLEV ETLYWKANGD RPFSVEYAND MPGSGFMPMK 180 ERKRTGEIVA GAAANVGETA WNMRGVAREN GSLLRFMKEE IPGVTSPMVY VAMMFSWFAW 240 HVEDHDLHSL NYLHLGAGKT WYGVPKDAAS AFEEVIRVYG FGEEINPLVT FAQLGEKTTV 300 MSPEVLINAG VPCCRLVQNA GEFVVTFPRA YHSGFSHGFN CGEASNIATP EWLRFAKDAA 360 IRRASINYPP MVSHFQLLYD LALAICSRAS IGNSAEPRSS RLKDKKKGEG EMLVKQMFVQ 420 DVIQNNELLY TLGQGSEVVL LPHNSSEIFV WSNLRVGSKY KVKPGLPFSL YSSEEAIKAS 480 DDIMLARDDR KQKAFSSVKT KSGGCMSTTP HEIQHSETEK GGGAAGDGFS DRGLFSCVKC 540 GIWTFACVAI VQPTESAAQY LMSADCNSFN DWIAGSGVSS HGMDPIDGEA NISDPNSFSG 600 SMEKHPPDGA YDIPVHSTDY HAQSISNTSK LKSNTAEIVE VGSHTETKRE SSALGLLAMT 660 YGNSSDSDED DVQPNSPVIS EDNLSGDGSW GARFHQDDSA SPVFEQGYDS GAERGPSQIS 720 SRSECEDEDS SQRSDFYEHC GHRRVNGDDN EYDSHNCSAK FTEEDILTSE QNYSPIADEH 780 DTAKISCAID PVGKPNLSFA HRCDEDSSRM HVFCLEHAVE VEKQLRPIGG VHILLLCHPD 840 YPNVEVEAKK AAEELEMDYG WKDIAFSMAT KEDEERIHMA LQSEESTPKN GDWAVKLGIN 900 LFYSAILSRS PLYNKQMPYN SIIYNAFGCT SPSKSSPEEA KVRGKGFGRQ KKLVMAGKWC 960 GKVWMSNQVH PLLLHRDPDE EERNFNACMK SDEKVGRKSE TSHKAQTTYT NRKVGKKRRS 1020 MPESTKVKKM KFEATEFADT DPEDSVDDKP EVETRRSVRK SKTQCRSIKR VKHQEEEEIL 1080 DGMSEDSQDE IKSLKSGRVL KLGTKQILKT KGRRSAAAIL SYSDDDSLDD RSHQHGLRSC 1140 NRKLKSTRMV RSQPKEIVDS ESEGSPSNES EIEEHQRRNI RSPGSSQRRV PVHRQFESVT 1200 DDDDDEMAGG PSTRLRKRIS RPIQERKEIK PTVKRQVQKP KGKKAAPLTK NIIPARKGPK 1260 MRLPVSRGPA MKGSKQELAL HKRNICPVKG CGKKFFSHKY LVQHRRVHMD DRPLKCPWKG 1320 CRMTFKWAWA RTEHIRVHTG ARPYVCNEPG CGQTFRFVSD FSRHKRKTGH AAKKARK |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6ip0_A | 3e-72 | 19 | 422 | 8 | 383 | Transcription factor jumonji (Jmj) family protein |
| 6ip4_A | 3e-72 | 19 | 422 | 8 | 383 | Arabidopsis JMJ13 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Histone demethylase that demethylates 'Lys-27' (H3K27me) of histone H3. Demethylates both tri- (H3K27me3) and di-methylated (H3K27me2) H3K27me (PubMed:21642989, PubMed:27111035). Demethylates also H3K4me3/2 and H3K36me3/2 in an in vitro assay (PubMed:20711170). Involved in the transcriptional regulation of hundreds of genes regulating developmental patterning and responses to various stimuli (PubMed:18467490). Binds DNA via its four zinc fingers in a sequence-specific manner, 5'-CTCTGYTY-3', to promote the demethylation of H3K27me3 and the regulation of organ boundary formation (PubMed:27111034, PubMed:27111035). Involved in the regulation of flowering time by repressing FLOWERING LOCUS C (FLC) expression (PubMed:15377760). Interacts with the NF-Y complexe to regulate SOC1 (PubMed:25105952). Mediates the recruitment of BRM to its target loci (PubMed:27111034). {ECO:0000269|PubMed:15377760, ECO:0000269|PubMed:18467490, ECO:0000269|PubMed:20711170, ECO:0000269|PubMed:21642989, ECO:0000269|PubMed:25105952, ECO:0000269|PubMed:27111034, ECO:0000269|PubMed:27111035}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00608 | ChIP-seq | Transfer from AT3G48430 | Download |
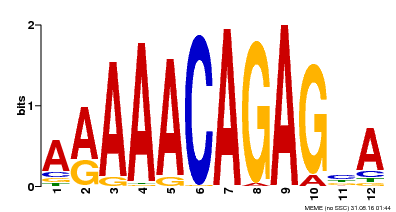
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021855215.1 | 0.0 | lysine-specific demethylase REF6 isoform X1 | ||||
| Swissprot | Q9STM3 | 0.0 | REF6_ARATH; Lysine-specific demethylase REF6 | ||||
| TrEMBL | A0A0K9QF77 | 0.0 | A0A0K9QF77_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010667395.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G48430.1 | 0.0 | relative of early flowering 6 | ||||



