 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sp_203670_kzwu.t1 | ||||||||
| Common Name | SOVF_203670 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Chenopodiaceae; Chenopodioideae; Anserineae; Spinacia
|
||||||||
| Family | Trihelix | ||||||||
| Protein Properties | Length: 490aa MW: 52425.2 Da PI: 9.6443 | ||||||||
| Description | Trihelix family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | trihelix | 49.7 | 9.5e-16 | 94 | 181 | 1 | 86 |
trihelix 1 rWtkqevlaLiearremeerlrrgklkkplWeevskkm....rergferspkqCkekwenlnkrykkikegekkrtsessstcpyfdqle 86
+W++ + +Li+a e+ +l+rg+lk+++W+ev++ + +++ ++ +qCk++++ ++k+yk +k+ ++ + s++p+fd+l+
Sp_203670_kzwu.t1 94 CWSEGATSVLIDAWGERYMELSRGNLKQKHWKEVADIVssreDYTKVPKTDIQCKNRIDTVKKKYKIEKQKVASG--GGPSRWPFFDRLD 181
7*************************************86555556677889*********************97..66778******98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF13837 | 1.5E-26 | 92 | 183 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009793 | Biological Process | embryo development ending in seed dormancy | ||||
| GO:0010431 | Biological Process | seed maturation | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 490 aa Download sequence Send to blast |
MDDEEEIRSQ PPSPESEPHS PPSGQITVTV AAAPPSSTTP SSTPPPSSLT LALPIQQFPR 60 PLAIISSGCG GGGSVGGGSS GGGGGGGGGG REDCWSEGAT SVLIDAWGER YMELSRGNLK 120 QKHWKEVADI VSSREDYTKV PKTDIQCKNR IDTVKKKYKI EKQKVASGGG PSRWPFFDRL 180 DRLIGPAATP TTSKGSGSGL GLGPMSTATV SVSTGGLSSH GKAVPVGIPV GVRPLPLPFS 240 SHSLQTPPHL QQWPQLKQQF QFSPVQQPVR YNSINSNSTS SPAAATPASN NVNRAPLEQQ 300 LQFHKHQQEL QFRRGEESDT DQEEWSHDDS GDSLPPERTG FDHRRKRPRM EVVNRGGGER 360 VKVKGKKSRE KKAAAVKVWG NSVRDLTRAI MKFGEAYESA ESAKLQHIVE MEMQRMKFTK 420 ELELQRMKFM MKTQLELSQL NGNGSNNSDS RGGGGGGGGS GGGGGGGGDS NHNHHLHENH 480 LNHANSDSSN |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 66 | 78 | SGCGGGGSVGGGS |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00356 | DAP | Transfer from AT3G14180 | Download |
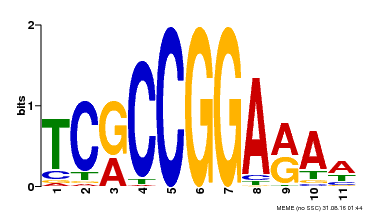
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021851201.1 | 0.0 | trihelix transcription factor ASIL2-like | ||||
| TrEMBL | A0A0K9Q9Q4 | 0.0 | A0A0K9Q9Q4_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010685259.1 | 1e-140 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G14180.1 | 2e-49 | sequence-specific DNA binding transcription factors | ||||



