 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sphfalx0010s0141.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Bryophyta; Sphagnophytina; Sphagnopsida; Sphagnales; Sphagnaceae; Sphagnum
|
||||||||
| Family | C3H | ||||||||
| Protein Properties | Length: 187aa MW: 20803.7 Da PI: 8.5014 | ||||||||
| Description | C3H family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-CCCH | 42.7 | 9.3e-14 | 106 | 132 | 1 | 27 |
--S---SGGGGTS--TTTTT-SS-SSS CS
zf-CCCH 1 yktelCrffartGtCkyGdrCkFaHgp 27
ykte+C+ +++ G+C+yG+rC+FaHg+
Sphfalx0010s0141.1.p 106 YKTEICNKWLEVGWCSYGSRCQFAHGM 132
9************************95 PP
| |||||||
| 2 | zf-CCCH | 20.3 | 9.9e-07 | 144 | 168 | 1 | 25 |
--S---SGGGGTS--TTTTT-SS-S CS
zf-CCCH 1 yktelCrffartGtCkyGdrCkFaH 25
yktelCr a++G C+y rC+F H
Sphfalx0010s0141.1.p 144 YKTELCRMVAAGGKCSYAHRCHFRH 168
9**********************88 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1000.10 | 3.7E-17 | 103 | 136 | IPR000571 | Zinc finger, CCCH-type |
| SuperFamily | SSF90229 | 2.75E-11 | 104 | 137 | IPR000571 | Zinc finger, CCCH-type |
| SMART | SM00356 | 7.4E-7 | 105 | 132 | IPR000571 | Zinc finger, CCCH-type |
| PROSITE profile | PS50103 | 16.305 | 105 | 133 | IPR000571 | Zinc finger, CCCH-type |
| Pfam | PF00642 | 8.8E-11 | 106 | 132 | IPR000571 | Zinc finger, CCCH-type |
| Gene3D | G3DSA:4.10.1000.10 | 1.8E-10 | 139 | 169 | IPR000571 | Zinc finger, CCCH-type |
| SuperFamily | SSF90229 | 1.96E-7 | 139 | 171 | IPR000571 | Zinc finger, CCCH-type |
| PROSITE profile | PS50103 | 12.258 | 143 | 171 | IPR000571 | Zinc finger, CCCH-type |
| SMART | SM00356 | 0.0011 | 143 | 170 | IPR000571 | Zinc finger, CCCH-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 187 aa Download sequence Send to blast |
MHPGGDARIL VPPVVPVQRV PHFAVPRSIA VRSNVDVMFA RRGPAASPLQ VHSPPGEFLP 60 PPSNRDERRA VGPNSTGPVD HGTSNGDIVE EAVLRLWDHV SDQGMYKTEI CNKWLEVGWC 120 SYGSRCQFAH GMEELRYVLR HPRYKTELCR MVAAGGKCSY AHRCHFRHTL TKEEKAKHFP 180 EATDGC* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1m9o_A | 2e-21 | 98 | 173 | 2 | 77 | Tristetraproline |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00383 | ampDAP | Transfer from AT1G68200 | Download |
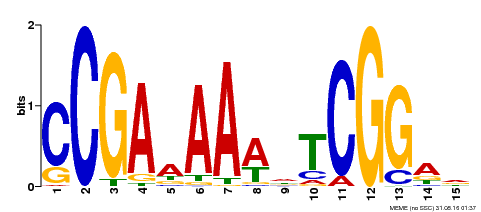
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021749707.1 | 5e-38 | zinc finger CCCH domain-containing protein 15-like | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G68200.1 | 1e-35 | C3H family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Sphfalx0010s0141.1.p |



