 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sphfalx0016s0059.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Bryophyta; Sphagnophytina; Sphagnopsida; Sphagnales; Sphagnaceae; Sphagnum
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 671aa MW: 70938.4 Da PI: 5.2311 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 63.3 | 5.1e-20 | 265 | 314 | 2 | 55 |
AP2 2 gykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
y+GVr+++ +g+WvAeIr p++ r+r +lg+f+taeeAa a+++a+ +l+g
Sphfalx0016s0059.1.p 265 LYRGVRQRH-WGKWVAEIRLPRN---RTRLWLGTFDTAEEAALAYDKAAYNLRG 314
59******9.**********955...5*************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 6.42E-30 | 264 | 324 | No hit | No description |
| PROSITE profile | PS51032 | 22.906 | 265 | 322 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 3.6E-22 | 265 | 323 | IPR016177 | DNA-binding domain |
| SMART | SM00380 | 1.7E-38 | 265 | 328 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 1.5E-31 | 265 | 322 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 7.3E-14 | 266 | 314 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.0E-10 | 266 | 277 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.0E-10 | 288 | 304 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009611 | Biological Process | response to wounding | ||||
| GO:0009736 | Biological Process | cytokinin-activated signaling pathway | ||||
| GO:0009873 | Biological Process | ethylene-activated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0045595 | Biological Process | regulation of cell differentiation | ||||
| GO:0071472 | Biological Process | cellular response to salt stress | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 671 aa Download sequence Send to blast |
MAVVADRLAY MPPTGHMNFP SCAAGSHVRQ ASAGSTEREF SMLGVQCDSN SWLADLGTSY 60 CSSSSSSSSS ERPGISNSDD GRADMRSCYS SFWPGFNYEV LLGSSGEAHE SSLSTTSSLQ 120 HLPVGQVVYS SAAPVDLRSV QQLVPVELSC SGSIQTQQPQ QLQRFDVVQQ LQQLQSMQIQ 180 QQQQQEQLLQ FQLLEAAAAS VYHENQLQWS SNRGRGSSEA GCSLGPRPIL MKKQQAHDKR 240 GVGVVGPSGS AAAAGGPRVQ QTSKLYRGVR QRHWGKWVAE IRLPRNRTRL WLGTFDTAEE 300 AALAYDKAAY NLRGDCARLN FPPQRNNSDD QLAASLSTGG STHSSDPNCS SWISSCSAGV 360 GTAATPGGLM SEKLPSTLSM KLETCIAYRK KAVQARNDEA GGGCSIQAAT PSPSLMEPDH 420 NRSGLQESNL ELAVSNDEPA ACMPSSSSSS LRVEGSCVTI ARALPAAGAD MSCSQIINTS 480 AAAAAAAQDL QGRSGGESAA GGTLMTQYRS SSQSSPSELV SDACSCIEDP AAGASDSCMW 540 EELVDILNNC DAAASEADLS SWADIIDFPL STRATSDSEP ELLQEPNPSL ICSKPEDAAA 600 AYSSNYHSSL SSSIVAWGSH HQQQLQEADN FAPGTAAAGN MKKKPLPARL VCSSSTLSPV 660 HQVRVWRNCN * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gcc_A | 7e-20 | 264 | 322 | 1 | 60 | ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 1 |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00026 | PBM | Transfer from AT1G78080 | Download |
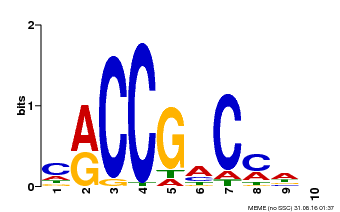
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G22190.1 | 5e-27 | ERF family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Sphfalx0016s0059.1.p |



