 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sphfalx0017s0148.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Bryophyta; Sphagnophytina; Sphagnopsida; Sphagnales; Sphagnaceae; Sphagnum
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 542aa MW: 58107.9 Da PI: 6.9883 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 56.8 | 5.1e-18 | 12 | 57 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+g+W++eEd l ++v+++G ++W++I++ ++ gR++k+c++rw +
Sphfalx0017s0148.1.p 12 KGPWSPEEDAALQRLVEKHGARNWSLISKGIP-GRSGKSCRLRWCNQ 57
79******************************.***********985 PP
| |||||||
| 2 | Myb_DNA-binding | 48.5 | 2e-15 | 66 | 108 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
++T+ Ed +++a++++G++ W+tIar ++ gRt++ +k++w++
Sphfalx0017s0148.1.p 66 PFTALEDSAIIQAHNLYGNK-WATIARILP-GRTDNAIKNHWNST 108
79******************.*********.***********986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 25.752 | 7 | 62 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.57E-31 | 9 | 105 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 6.1E-17 | 11 | 60 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 6.0E-18 | 12 | 57 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.1E-23 | 13 | 65 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.27E-15 | 14 | 56 | No hit | No description |
| SMART | SM00717 | 2.4E-14 | 63 | 111 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS51294 | 19.326 | 63 | 113 | IPR017930 | Myb domain |
| CDD | cd00167 | 3.60E-11 | 66 | 109 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 5.8E-20 | 66 | 112 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 1.0E-12 | 66 | 108 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 542 aa Download sequence Send to blast |
MAALVEEFDR IKGPWSPEED AALQRLVEKH GARNWSLISK GIPGRSGKSC RLRWCNQLSP 60 LVHHRPFTAL EDSAIIQAHN LYGNKWATIA RILPGRTDNA IKNHWNSTLR RRHLADKTSQ 120 RSTSRGGAGD NRISPAAEEE EEDEEEIGSS FDGRKRNYSN EISSTDRSVQ QEESGCWEVD 180 SLRLKKLSFG AGASDDDDHV QLQQHMKNKR PISFGPDDDD PAAASHHRLK KKQLLASGCE 240 DASTVVSSAL LASDDDDVQM KLKKKLSFGG SDSSTQLMPA PALFRPVARA SAFNTFNPTP 300 PAPPSYTTHA SSSSSSGVVQ NSSTSIERHH QQEIHHQANK ASFGVNAVGA AGLLPMDPPT 360 SLSLSLPGTI IQSSISQAAS EQVAVLEKQP SMSSFTKGSS VAEVPAEMYD LIGATDKEAC 420 ANLMSLAVKS VVAQALAPVF PQQGQAAAGT KWISTSATAA PLGSLDAALN AGLLAIMRDM 480 VAKEVHNYLA AVHSSSCLPF FHALATTHHE HAASLASRDP TFSNQPEFLG FMATTAPRKA 540 V* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1mse_C | 1e-36 | 11 | 112 | 3 | 104 | C-Myb DNA-Binding Domain |
| 1msf_C | 1e-36 | 11 | 112 | 3 | 104 | C-Myb DNA-Binding Domain |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00632 | PBM | Transfer from PK28565.1 | Download |
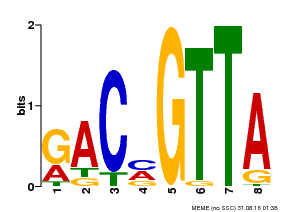
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G23290.1 | 8e-56 | myb domain protein 70 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Sphfalx0017s0148.1.p |



