 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sphfalx0019s0143.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Bryophyta; Sphagnophytina; Sphagnopsida; Sphagnales; Sphagnaceae; Sphagnum
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 535aa MW: 57609.4 Da PI: 5.1102 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 53.8 | 4.6e-17 | 16 | 63 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT Ed +lv +v+++G g+W+++ ++ g+ R++k+c++rw ++l
Sphfalx0019s0143.1.p 16 KGPWTSTEDAILVSYVNKHGEGNWNSVQKHSGLYRCGKSCRLRWANHL 63
79******************************************9986 PP
| |||||||
| 2 | Myb_DNA-binding | 48.7 | 1.7e-15 | 69 | 112 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
+g++T++E+ +v+++ +lG++ W++ a+ ++ gRt++++k++w++
Sphfalx0019s0143.1.p 69 KGAFTPDEERTIVELHSKLGNK-WARMAAQLP-GRTDNEIKNYWNT 112
799*******************.*********.***********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 15.812 | 11 | 63 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 7.49E-30 | 13 | 110 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.5E-14 | 15 | 65 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 4.3E-15 | 16 | 63 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.4E-23 | 17 | 70 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 5.60E-11 | 18 | 63 | No hit | No description |
| PROSITE profile | PS51294 | 24.666 | 64 | 118 | IPR017930 | Myb domain |
| SMART | SM00717 | 8.7E-15 | 68 | 116 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.5E-14 | 69 | 112 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 5.3E-25 | 71 | 118 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.80E-10 | 71 | 112 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009789 | Biological Process | positive regulation of abscisic acid-activated signaling pathway | ||||
| GO:0043068 | Biological Process | positive regulation of programmed cell death | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0045926 | Biological Process | negative regulation of growth | ||||
| GO:0048235 | Biological Process | pollen sperm cell differentiation | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 535 aa Download sequence Send to blast |
MDLPQEIFAD AGNLKKGPWT STEDAILVSY VNKHGEGNWN SVQKHSGLYR CGKSCRLRWA 60 NHLRPNLKKG AFTPDEERTI VELHSKLGNK WARMAAQLPG RTDNEIKNYW NTRIKRRMRA 120 GLPVYPMDIP KSAASLSQFY FKQEDDGRKM GTRADSESEF ASSVVTLLGD RQSHQLSSLH 180 GGHQARSFYS IMSITDLPLT SVVANQGSAG VGKGIVNPSL PHIKRVQHSS PYGGTDGSHG 240 SVSFSQLSDG SSKTFQPELC TEASRTCTHN STRRPMMSVF ADGLDPYTVP YDGTCGLISM 300 PFGSFTPNAS SKLELPLSQS AESADSAGTP RSAITSPLMS LSRSSQLLSE DDSFGSNASD 360 FLDALMKEAH PTEGFDQVRT DLIDQLLALT AGNISPDVAA LLVGTDKTRW GENMNPMATI 420 EAHASVYGEG MSQNCCPGSL KEPGTQCHLL SGEVLSLSAS QVAGPRVEDI PAEIPLGGCT 480 DEDFLTLLDF ASADAVPEWY TPAEYFSAGG IPCTSLVDVI DNVPSPEVAV DLDY* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 2e-32 | 13 | 118 | 4 | 108 | B-MYB |
| 1h8a_C | 3e-32 | 14 | 118 | 25 | 128 | MYB TRANSFORMING PROTEIN |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00490 | DAP | Transfer from AT5G06100 | Download |
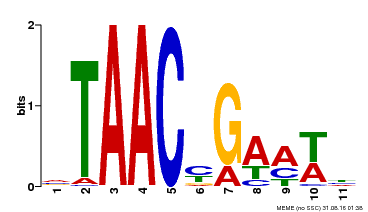
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024397297.1 | 1e-158 | transcription factor GAMYB-like | ||||
| Refseq | XP_024397298.1 | 1e-158 | transcription factor GAMYB-like | ||||
| TrEMBL | A0A2K1JEJ0 | 1e-157 | A0A2K1JEJ0_PHYPA; Uncharacterized protein | ||||
| STRING | PP1S66_200V6.1 | 1e-157 | (Physcomitrella patens) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G06100.1 | 1e-70 | myb domain protein 33 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Sphfalx0019s0143.1.p |



