 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sphfalx0028s0189.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Bryophyta; Sphagnophytina; Sphagnopsida; Sphagnales; Sphagnaceae; Sphagnum
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 621aa MW: 67569 Da PI: 5.8776 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 54.9 | 2.3e-17 | 50 | 100 | 2 | 56 |
AP2 2 gykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkklege 56
+y+GVr+++ +gr++AeIrdp++ + r +lg+f+tae+Aa a++ a+++++g+
Sphfalx0028s0189.1.p 50 RYRGVRRRP-WGRYAAEIRDPNT---KERKWLGTFDTAEDAAIAYDWAARSMRGA 100
69*******.**********933...5*************************995 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51032 | 21.733 | 50 | 107 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 6.5E-29 | 50 | 106 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 1.18E-20 | 50 | 107 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 1.9E-10 | 50 | 99 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 3.1E-32 | 50 | 113 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 4.06E-28 | 50 | 106 | No hit | No description |
| PRINTS | PR00367 | 5.0E-9 | 51 | 62 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 5.0E-9 | 73 | 89 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010102 | Biological Process | lateral root morphogenesis | ||||
| GO:0010432 | Biological Process | bract development | ||||
| GO:0010451 | Biological Process | floral meristem growth | ||||
| GO:0010582 | Biological Process | floral meristem determinacy | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 621 aa Download sequence Send to blast |
MVGRIDAFAQ RQLLPQQSHC EPGLAVTRGS SMRKGGLETK KGRRSREPGR YRGVRRRPWG 60 RYAAEIRDPN TKERKWLGTF DTAEDAAIAY DWAARSMRGA KARTNFVYPT HHTCIMSTTL 120 ATDRNSRAAG DDHSYRDQHH QLQQLSGSEA PNNNFCAVNV GAPASKPRKE DWTNTGFHDS 180 QTSSRSDAYE SIYQSVEKLA QAISDIGPAT RPRPASSSSL LLPHTDPRGQ DSINGACMRE 240 EGNESVVGRK QWLQSEYTYE CSLYESASRA PKSNLTPPIL QVQIPDFCEE PPGGSCAGLL 300 SPPCNSTMAP LSHGAAALSH QSSLSVPLTT SSEVPPHVRK YSEEVDTLAQ VIAQSSVDFN 360 RPMITAGKPP QQQQHEQIPP ASHRNCMVST QVSYSEDTTP SLPADSLSTP DSPGLFSLSS 420 EAESYGPSPT AAEVSSPLCS SGSGIVTSPS RNPDNLQSIL PANLSCPAVE RWSQPQSRDW 480 SWGEPAAAAV QIPQVLQTAC TLAAAWQCSN SPMEEVKCEP ASSSAASWQT LCALPAAAAA 540 WAQTPRASCE LVGTRKDDWQ DYNIKRAEQQ QQFSGLLGDL VGESFTTNDT LYMESIPFPE 600 HPLDDCHLFP FLLDECRIFV * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5wx9_A | 5e-20 | 47 | 106 | 11 | 71 | Ethylene-responsive transcription factor ERF096 |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 39 | 57 | KKGRRSREPGRYRGVRRRP |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00520 | DAP | Transfer from AT5G18560 | Download |
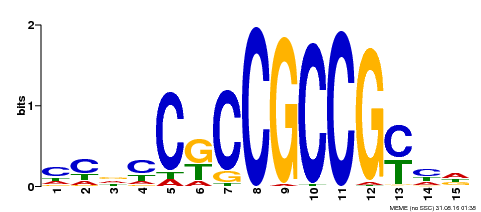
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G13910.1 | 3e-14 | ERF family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Sphfalx0028s0189.1.p |



