 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sphfalx0034s0063.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Bryophyta; Sphagnophytina; Sphagnopsida; Sphagnales; Sphagnaceae; Sphagnum
|
||||||||
| Family | ARR-B | ||||||||
| Protein Properties | Length: 829aa MW: 88995.6 Da PI: 6.2851 | ||||||||
| Description | ARR-B family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 90.6 | 1.4e-28 | 297 | 350 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
kpr++W+ eLH++Fv+av+qL G++kA+Pk+ilelm+v+gLt+e+v+SHLQkYRl
Sphfalx0034s0063.1.p 297 KPRVVWSVELHQQFVSAVNQL-GIDKAVPKKILELMSVHGLTRENVASHLQKYRL 350
79*******************.********************************8 PP
| |||||||
| 2 | Response_reg | 78.6 | 2.1e-26 | 110 | 218 | 1 | 109 |
EEEESSSHHHHHHHHHHHHHTTCEEEEEESSHHHHHHHHHHHH..ESEEEEESSCTTSEHHHHHHHHHHHTTTSEEEEEESTTTHHHHH CS
Response_reg 1 vlivdDeplvrellrqalekegyeevaeaddgeealellkekd..pDlillDiempgmdGlellkeireeepklpiivvtahgeeedal 87
vl+vdD+p+ +++l ++l++ y +v+++ +++alell+e + +Dl++ D+ mp+mdG++ll+ e++lp+i++++ ge + +
Sphfalx0034s0063.1.p 110 VLVVDDDPICLMILDRMLRQCSY-QVTTCGRATKALELLREVKdkFDLVISDVYMPDMDGFKLLELVGL-EMDLPVIMMSSDGETSAVM 196
89*********************.***************776668*******************87754.558**************** PP
HHHHTTESEEEESS--HHHHHH CS
Response_reg 88 ealkaGakdflsKpfdpeelvk 109
+ + Ga d+l Kp+ eel++
Sphfalx0034s0063.1.p 197 KGITHGACDYLLKPVRLEELRN 218
********************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF52172 | 1.99E-36 | 107 | 232 | IPR011006 | CheY-like superfamily |
| Gene3D | G3DSA:3.40.50.2300 | 2.0E-43 | 107 | 244 | No hit | No description |
| SMART | SM00448 | 1.7E-31 | 108 | 220 | IPR001789 | Signal transduction response regulator, receiver domain |
| PROSITE profile | PS50110 | 44.42 | 109 | 224 | IPR001789 | Signal transduction response regulator, receiver domain |
| Pfam | PF00072 | 2.7E-23 | 110 | 219 | IPR001789 | Signal transduction response regulator, receiver domain |
| CDD | cd00156 | 2.32E-28 | 111 | 223 | No hit | No description |
| SuperFamily | SSF46689 | 1.76E-19 | 294 | 354 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 11.509 | 294 | 353 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.7E-29 | 296 | 355 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.3E-24 | 297 | 350 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 4.7E-7 | 299 | 349 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009873 | Biological Process | ethylene-activated signaling pathway | ||||
| GO:0010082 | Biological Process | regulation of root meristem growth | ||||
| GO:0010119 | Biological Process | regulation of stomatal movement | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0071368 | Biological Process | cellular response to cytokinin stimulus | ||||
| GO:0080113 | Biological Process | regulation of seed growth | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 829 aa Download sequence Send to blast |
MHHSPIAVCT TMVGEELSRS RVGVGDLGSA MASLQQQQPL VVMSGGQQQH QQHQQQQCGS 60 ATNSSHNATS THGIITTNLH HNNNTTTRAS DGFGKMDNPA VDEFPAGLRV LVVDDDPICL 120 MILDRMLRQC SYQVTTCGRA TKALELLREV KDKFDLVISD VYMPDMDGFK LLELVGLEMD 180 LPVIMMSSDG ETSAVMKGIT HGACDYLLKP VRLEELRNIW QHVVRKKRNE AKDVEHSTSH 240 EDAERHKRGG AGDDADYTSS ATDTTDGNCK LTKRKKEFKE EDDDNEQEND DPSTLKKPRV 300 VWSVELHQQF VSAVNQLGID KAVPKKILEL MSVHGLTREN VASHLQKYRL YLRRLSGVTS 360 QQNGMNMTFG GSETFGNLSS LDGISDLRTL AASGQLSPQT LAALHANMIS RVGMPNGMGL 420 PSSLDPSVLA HALQNVPLPR PQMDGALLAN QSGMAQSIPG PPVLDVDPHL QPHHLSAMGQ 480 LSQLDDMPGL RALQHQLAGA CASNSNGIGL NNPLGGSSGN LVASSNNESL MMQLLQQRVQ 540 QQQQGGGLSV NVPQPSAVLG SPRLLSTDMN LGQAGSLTNM GRGPGSSSLR MPGGASVPHS 600 LGSLFRPTGS GVQALNPVCS SGGSFSGSTT VLSPRSGGVP GCPPATDDMA RSQHSMRTSL 660 NPLGQATRGH EQVLSQSSRP TWLGSQGGMI SGEHGQSIES GLSQFPSQSQ THASGFAGNV 720 RQQSQPEYTS RGMNFPVRLG LDVRPLFDGS SQSHSEQQPT TDIEQKLPKD ERASDNVVIT 780 KLVDGGMVSH DQSEDLLNFF FKQSHEGMSF PDSELASDGY TMDNFYVK* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1irz_A | 1e-21 | 296 | 356 | 4 | 64 | ARR10-B |
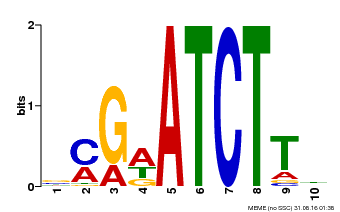
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00054 | PBM | Transfer from AT4G16110 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024363770.1 | 1e-180 | two-component response regulator ARR12-like | ||||
| TrEMBL | A0A2K1IGK7 | 1e-178 | A0A2K1IGK7_PHYPA; Uncharacterized protein | ||||
| STRING | PP1S10_278V6.1 | 1e-176 | (Physcomitrella patens) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16110.1 | 1e-114 | response regulator 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Sphfalx0034s0063.1.p |



