 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sphfalx0034s0088.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Bryophyta; Sphagnophytina; Sphagnopsida; Sphagnales; Sphagnaceae; Sphagnum
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 430aa MW: 44657.1 Da PI: 7.4812 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 170.7 | 8e-53 | 26 | 123 | 2 | 96 |
DUF822 2 gsgrkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpl.eeae.aagssasa 88
++ r ptwkErEnnkrRERrRRaiaaki+aGLR++Gnyklpk++DnneVlkALc+eAGw+ve+DG tyrkg+kp+ e+++ + + a +
Sphfalx0034s0088.1.p 26 SGSRLPTWKERENNKRRERRRRAIAAKIFAGLRSYGNYKLPKHCDNNEVLKALCAEAGWTVEEDGSTYRKGTKPPvEQMQeVCTTPALV 114
789************************************************************************95555055566669 PP
DUF822 89 spesslq.s 96
sp+ss+
Sphfalx0034s0088.1.p 115 SPTSSYPgA 123
999999833 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 4.7E-51 | 27 | 129 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 430 aa Download sequence Send to blast |
MVAARDHQRV DAPGAAAERG DIIMTSGSRL PTWKERENNK RRERRRRAIA AKIFAGLRSY 60 GNYKLPKHCD NNEVLKALCA EAGWTVEEDG STYRKGTKPP VEQMQEVCTT PALVSPTSSY 120 PGASDGTSLI PWLKGLSSGR GSGTASTSSS AGLPTLHLVH AGTSSCAPVT PPLSSPTAKG 180 IPVKPDWDAI AKDGLPECPT HAFSQAHVNA WSAVHHPSFL AAAAAAAASN QSPLCPGSYC 240 ETPDGCRTPA ADEVESDLSP TTALEFANVC SSSSTKWANG IRVRTAGSSA AAGAILGLAA 300 GPPGSAVSGC HVQLEGMMGL GPFPAPAAVQ SDSPETPYPS TWRIPISLQS SAANTPVPAR 360 TVPRLLNSDK VENPDAVVHV PWGEAAAAAE DHVAMQVVSR RKLAVQPDEL ELTLGSSSSR 420 GRSPNLQCQ* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5zd4_A | 1e-21 | 29 | 99 | 372 | 442 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_B | 1e-21 | 29 | 99 | 372 | 442 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_C | 1e-21 | 29 | 99 | 372 | 442 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_D | 1e-21 | 29 | 99 | 372 | 442 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00248 | DAP | Transfer from AT1G78700 | Download |
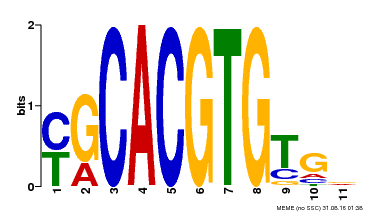
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024357987.1 | 1e-106 | BES1/BZR1 homolog protein 4-like | ||||
| TrEMBL | A0A2K1ITH7 | 1e-105 | A0A2K1ITH7_PHYPA; Uncharacterized protein | ||||
| STRING | PP1S335_68V6.1 | 1e-104 | (Physcomitrella patens) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G78700.1 | 2e-35 | BES1/BZR1 homolog 4 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Sphfalx0034s0088.1.p |



