 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sphfalx0066s0025.6.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Bryophyta; Sphagnophytina; Sphagnopsida; Sphagnales; Sphagnaceae; Sphagnum
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 467aa MW: 52176.7 Da PI: 7.9492 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 34.5 | 4.3e-11 | 184 | 224 | 4 | 48 |
XCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 4 lkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkke 48
+k+ rr+++NReAAr+sR+RKka++++Le +L + + e
Sphfalx0066s0025.6.p 184 SKALRRLAQNREAARKSRLRKKAYVQQLESSRIKLNQLEQ----E 224
7999*************************86555544443....3 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.20.5.170 | 5.5E-8 | 178 | 230 | No hit | No description |
| SMART | SM00338 | 3.4E-6 | 181 | 244 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 9.174 | 183 | 227 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 1.6E-7 | 185 | 225 | IPR004827 | Basic-leucine zipper domain |
| CDD | cd14708 | 1.15E-22 | 185 | 231 | No hit | No description |
| SuperFamily | SSF57959 | 8.75E-7 | 186 | 230 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 188 | 203 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF14144 | 3.6E-34 | 266 | 341 | IPR025422 | Transcription factor TGA like domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009410 | Biological Process | response to xenobiotic stimulus | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005737 | Cellular Component | cytoplasm | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 467 aa Download sequence Send to blast |
MISVEAAKVL WYTILHMCPF HLEMMPCCGG WRMTSIKSNF SKESVEAAKV LWYTALCTIC 60 PIHLVMMPCY GGWSTISVKN NFSKESVEAA KQQQQYNIPK QVTTQLGVQA HFLYCFYSIE 120 PIGFTAISTQ KLFEHQADSW MADNSPRTDT STDLEADAKL DDGHHGTTGV STASDHETTK 180 NGDSKALRRL AQNREAARKS RLRKKAYVQQ LESSRIKLNQ LEQELQRARQ QNPHSASANG 240 VNVAINPGAT AFDMEYARWV EEQQRQMCEL RAALQAHVAD NELRLLVDGG MAHYDDIFRL 300 KAVAAKADVF HLVSGMWKTP AERCFMWMGG FRPSELLKIL IPQIEPLTEQ QLLGICNLQQ 360 SSQQAEDALS QGMEALQQSL ADIVAGGLLG SSPNVANYMG QMAMAMGKLG TLESFVRQAD 420 HLRQQTLQQM HRILTTRQAA RGLLAMGDYF ARLRALSSLW SARPRD* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
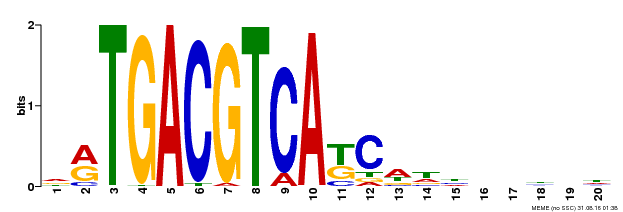
| UniProt | Transcriptional activator that binds specifically to the DNA sequence 5'-TGACG-3'. Recognizes ocs elements like the as-1 motif of the cauliflower mosaic virus 35S promoter. Binding to the as-1-like cis elements mediate auxin- and salicylic acid-inducible transcription. Binds to the hexamer motif 5'-ACGTCA-3' of histone gene promoters. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00492 | DAP | Transfer from AT5G06950 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024379754.1 | 0.0 | transcription factor TGA6-like isoform X2 | ||||
| Swissprot | Q41558 | 1e-154 | HBP1C_WHEAT; Transcription factor HBP-1b(c1) (Fragment) | ||||
| TrEMBL | A0A2R6XAK6 | 0.0 | A0A2R6XAK6_MARPO; Uncharacterized protein | ||||
| TrEMBL | A0A2R6XAL5 | 0.0 | A0A2R6XAL5_MARPO; Uncharacterized protein | ||||
| STRING | PP1S258_7V6.1 | 1e-179 | (Physcomitrella patens) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G12250.2 | 1e-135 | TGACG motif-binding factor 6 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Sphfalx0066s0025.6.p |



