 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sphfalx0066s0104.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Bryophyta; Sphagnophytina; Sphagnopsida; Sphagnales; Sphagnaceae; Sphagnum
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 806aa MW: 84842.6 Da PI: 6.6908 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 53.9 | 4.1e-17 | 14 | 61 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g W++eEde+lv+++++ G g+W+++++ g+ R++k+c++rw +yl
Sphfalx0066s0104.1.p 14 KGLWSPEEDEKLVRYITKCGHGCWSAVPKQAGLQRCGKSCRLRWINYL 61
678*******************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 45 | 2.5e-14 | 67 | 110 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
rg +++ E+ l+vd+++ lG++ W+ Ia ++ gRt++++k++w++
Sphfalx0066s0104.1.p 67 RGGFSPAEENLIVDLHAVLGNR-WSQIATQLP-GRTDNEIKNFWNS 110
7889******************.*********.***********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 2.5E-26 | 6 | 64 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 24.703 | 9 | 65 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.99E-28 | 11 | 108 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 3.5E-13 | 13 | 63 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 5.7E-15 | 14 | 61 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 3.80E-11 | 17 | 61 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 5.6E-24 | 65 | 116 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 19.085 | 66 | 116 | IPR017930 | Myb domain |
| SMART | SM00717 | 4.0E-13 | 66 | 114 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.7E-12 | 67 | 110 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.43E-9 | 70 | 109 | No hit | No description |
| PROSITE pattern | PS00141 | 0 | 179 | 190 | IPR001969 | Aspartic peptidase, active site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0001944 | Biological Process | vasculature development | ||||
| GO:0006508 | Biological Process | proteolysis | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0010089 | Biological Process | xylem development | ||||
| GO:0010119 | Biological Process | regulation of stomatal movement | ||||
| GO:0010214 | Biological Process | seed coat development | ||||
| GO:0048364 | Biological Process | root development | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0004190 | Molecular Function | aspartic-type endopeptidase activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 806 aa Download sequence Send to blast |
MGRHSCCHKQ KLKKGLWSPE EDEKLVRYIT KCGHGCWSAV PKQAGLQRCG KSCRLRWINY 60 LRPDLKRGGF SPAEENLIVD LHAVLGNRWS QIATQLPGRT DNEIKNFWNS CIKKRLRQMG 120 IDPNTHNPVT AGAGGVTSSA SHVTSSNPAA AGNCHATASS NNMMILPGLH MSSYNNNKAL 180 VDTGSSLLTM QSSSTIGGNF NGRFQGGSNH EVLIKAAASQ SHSSRLQLMN QAQAAGNNNY 240 HHRHHHGSSM MMEPDPHHML LGPLAAAARN SQVVEMMNTA GGRKFENVEF NREGSRSAAA 300 AAAAAALNFQ GMNNTFQASN TSCSSSGLST DNLLERLMLS AKQQSSGSMQ QYLPPKVPGL 360 LQGFEAMRNC SSVKLMQSNC RELAAVAGHD DETAASAGRS SSSSVIKAES MTQCGGPTTM 420 VPNLYNPVFW LFQGSAAGNA AAAASSQADE SSVASLGTRP AGAHGLDEML PPASSKQAGG 480 ITSANAGDSA LSAAAQSGSR VCQDSTHDMK AFQRQFHGQH GPAENFYPSA VEQDHGDLSS 540 ITASITQPGN AGSGRMIPSH QLQLLLSSML EKPTSAGADE LLHHDFNAAS AGMQQNNYNE 600 MSSGQADGHG IQAAAGAHNQ QWDSSVSSST SNNNNNREGT PGTTTTTQQS GLFESNLMWA 660 NFSEKAGAAG DHPPQQNQVA AAAAATSDIM SQGEQSDQIS AAAVSPGDSD GDDTVMQWCS 720 DLTTLPGSPE DDDHHVQRSP LAPSTTASNH GPSSHAPPPA IISAWQLHQA HHGAPDLYDA 780 REGPVVATLC MSQELQRMAA VLDEI* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 1e-27 | 12 | 116 | 5 | 108 | B-MYB |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00134 | DAP | Transfer from AT1G09540 | Download |
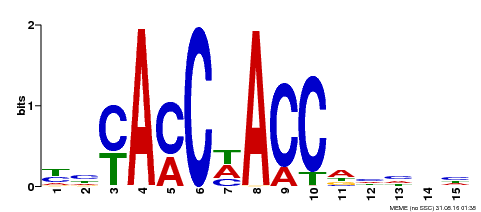
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09540.1 | 2e-79 | myb domain protein 61 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Sphfalx0066s0104.1.p |



