 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sphfalx0082s0020.2.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Bryophyta; Sphagnophytina; Sphagnopsida; Sphagnales; Sphagnaceae; Sphagnum
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 1038aa MW: 112170 Da PI: 5.8781 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 52.7 | 1e-16 | 60 | 106 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g WT+eEde l +av++l g++Wk+Ia+ + Rt+ qc +rwqk+l
Sphfalx0082s0020.2.p 60 KGGWTPEEDEVLRRAVQLLKGKNWKKIAEFFT-DRTDVQCLHRWQKVL 106
688****************************9.************986 PP
| |||||||
| 2 | Myb_DNA-binding | 54.3 | 3.2e-17 | 112 | 158 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+ Ed+l+ ++v+++G+ W+ Ia+ ++ gR +kqc++rw+++l
Sphfalx0082s0020.2.p 112 KGAWTKVEDDLIMELVAKHGPTKWSMIAQSLP-GRIGKQCRERWHNHL 158
79******************************.*************97 PP
| |||||||
| 3 | Myb_DNA-binding | 56.3 | 7.2e-18 | 164 | 208 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r +W+++Ed+ l++a++++G++ W+ Ia+ ++ gRt++++k++w++
Sphfalx0082s0020.2.p 164 REAWSEQEDLTLINAHQLYGNK-WAEIAKFLP-GRTDNSIKNHWHST 208
789*******************.*********.***********975 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 17.205 | 55 | 106 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 2.37E-15 | 57 | 112 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.5E-14 | 59 | 108 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 9.6E-15 | 60 | 106 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 9.9E-21 | 62 | 118 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 5.87E-13 | 63 | 106 | No hit | No description |
| PROSITE profile | PS51294 | 30.569 | 107 | 162 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 8.99E-31 | 109 | 205 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.4E-15 | 111 | 160 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.7E-16 | 112 | 158 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 8.68E-14 | 114 | 158 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 7.0E-26 | 119 | 165 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 23.135 | 163 | 213 | IPR017930 | Myb domain |
| SMART | SM00717 | 2.7E-17 | 163 | 211 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.1E-16 | 164 | 207 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.1E-23 | 166 | 212 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 7.61E-13 | 166 | 206 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0032875 | Biological Process | regulation of DNA endoreduplication | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003713 | Molecular Function | transcription coactivator activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1038 aa Download sequence Send to blast |
MASTQGNNNN NSCTTTSTEH MQQQRSSACS SGLESDDDTS RDHLPPVHGR TGGPTRRSSK 60 GGWTPEEDEV LRRAVQLLKG KNWKKIAEFF TDRTDVQCLH RWQKVLNPDL VKGAWTKVED 120 DLIMELVAKH GPTKWSMIAQ SLPGRIGKQC RERWHNHLNP NIKREAWSEQ EDLTLINAHQ 180 LYGNKWAEIA KFLPGRTDNS IKNHWHSTMK KRVDPETATD PVSKALAAYQ AQQDAITSAG 240 SSMHLEPAMN MTGRVGPSTS KFKTFCQPFE CFRGMSTNPY MTFYDGDGDL PMSKAAVTAA 300 AAAPLQQSSL HAWLESGALS TTMDLPRHPS FSLVSPMSPS SKGFPSMAIP SPLSTSASFL 360 TSSMPVAPLD SMDSITPLHG RPAATSGQSV ATMDTQQPET KSMMEGVTKL LDTNPTSAAE 420 CDDTQGDNPT GNEVVGYQDE VLRAANQAVG DYHPGATYML MEVELEEEDR DMEGSMGIEM 480 EGLFYEPPRF PCSSESPFVS YDLIQSGQAL QAYSPLGIRQ MIVSMGNCIT PPPYHLSSES 540 PFQGNSPQSR LRSAAQSFSG SPSILRKRPR HLVIPPKQGN NNTGGKKDRR LGPSWMSPRV 600 KEEKTTTADL LNCSVKTPDS TVQCSSLREE SVKPDCQART ALFVSPPYHL GKSSSSEVVP 660 GDLLNSEYRT QDCTKDRSGG VGCYSSLPTS ARDGREVAAA CSTGISEAAG GKCTKQQSHG 720 LTHEYHTGSI QWADSEQATK RTNGKGKPFF PLEANTLSTH VLVGTEKDDS GMVLGGNPGF 780 GEMRASCETS MLRSESLIAA AAVSSKAPDQ TEMMSQGGSM PPLGVWASLR YASSPGSAVL 840 QEAPGIDWLA ELESINFASV MSDGSFSSPT AIWREHWRVD PFPSAEKDGA ISSVEGSDAP 900 ARGREDDALG IMEHLNEDNA AAYCEAEEVL ASLPSAVCMA SPHSQYRTRT NVGHSPHDCK 960 ENMRNISVDG GTSSSFVNWL PPFHVDFLSP DRGITRTPVR GGSTLGLNGG LDETHAVGGG 1020 GSCDQFSPSI YLLKECR* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h88_C | 4e-66 | 60 | 213 | 6 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1h89_C | 4e-66 | 60 | 213 | 6 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00505 | DAP | Transfer from AT5G11510 | Download |
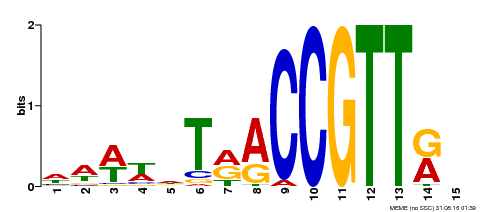
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| STRING | PP1S120_159V6.1 | 1e-122 | (Physcomitrella patens) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G11510.2 | 1e-87 | myb domain protein 3r-4 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Sphfalx0082s0020.2.p |



