 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sphfalx0095s0033.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Bryophyta; Sphagnophytina; Sphagnopsida; Sphagnales; Sphagnaceae; Sphagnum
|
||||||||
| Family | ARF | ||||||||
| Protein Properties | Length: 827aa MW: 90864.5 Da PI: 6.3595 | ||||||||
| Description | ARF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 75.9 | 4.7e-24 | 166 | 266 | 1 | 98 |
EEEE-..-HHHHTT-EE--HHH.HTT......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEE CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFk 83
f+k+lt sd++++g +++p++ aee+ ++++++ +l+ +d +g +W++++iyr++++r++lt+GW+ Fv++++L +gD+v+F
Sphfalx0095s0033.1.p 166 FCKTLTASDTSTHGGFSVPRRAAEEClpeldyTMNPPCQ-ELVAKDVHGVEWKFRHIYRGHPRRHLLTTGWSVFVSSKRLVAGDSVLFL 253
99*****************************97777785.9************************************************ PP
E-SSSEE..EEEEE- CS
B3 84 ldgrsefelvvkvfr 98
+ ++++l+++v+r
Sphfalx0095s0033.1.p 254 R--GENEQLRLGVRR 266
3..367777999988 PP
| |||||||
| 2 | Auxin_resp | 115.2 | 5.6e-38 | 292 | 374 | 1 | 83 |
Auxin_resp 1 aahaastksvFevvYnPrastseFvvkvekvekalkvkvsvGmRfkmafetedsserrlsGtvvgvsdldpvrWpnSkWrsLk 83
aahaa++k F+v+Y+P++++seFv++++k++ka++++vsvGmRfkm+fe+eds err +Gt++g+ d+dpvrWpnS WrsLk
Sphfalx0095s0033.1.p 292 AAHAATEKLRFSVIYHPSTCSSEFVIPYHKYMKAINSSVSVGMRFKMRFESEDSTERRHAGTITGIGDVDPVRWPNSYWRSLK 374
79*******************************************************************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.40.330.10 | 5.0E-40 | 158 | 280 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 2.88E-41 | 162 | 295 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 1.08E-22 | 165 | 266 | No hit | No description |
| Pfam | PF02362 | 6.3E-22 | 166 | 266 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 12.351 | 166 | 268 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 1.3E-23 | 166 | 268 | IPR003340 | B3 DNA binding domain |
| Pfam | PF06507 | 3.0E-34 | 292 | 374 | IPR010525 | Auxin response factor |
| Pfam | PF02309 | 1.5E-5 | 718 | 754 | IPR033389 | AUX/IAA domain |
| SuperFamily | SSF54277 | 1.52E-10 | 719 | 797 | No hit | No description |
| PROSITE profile | PS51745 | 29.822 | 719 | 802 | IPR000270 | PB1 domain |
| Pfam | PF02309 | 6.7E-7 | 765 | 798 | IPR033389 | AUX/IAA domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0008285 | Biological Process | negative regulation of cell proliferation | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009911 | Biological Process | positive regulation of flower development | ||||
| GO:0010047 | Biological Process | fruit dehiscence | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0010227 | Biological Process | floral organ abscission | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0048481 | Biological Process | plant ovule development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 827 aa Download sequence Send to blast |
MMSESNFRVP AAAAAATMRS CNGGGSGVVA LQQHISPPMS QCPLPSSSDS SPVGIPGNED 60 LDSELWHACA GPLISLPPVN SLVVYWPQGH LEQVANCTPQ AAVQPPIPQY NLPPHILCRL 120 TDITLMAHPD TDKVYAQMDL TPVRESEKPL VAEEPPPAVE RKSNSFCKTL TASDTSTHGG 180 FSVPRRAAEE CLPELDYTMN PPCQELVAKD VHGVEWKFRH IYRGHPRRHL LTTGWSVFVS 240 SKRLVAGDSV LFLRGENEQL RLGVRRALKQ QQDAPSMVVS NQSTHIGVLA TAAHAATEKL 300 RFSVIYHPST CSSEFVIPYH KYMKAINSSV SVGMRFKMRF ESEDSTERRH AGTITGIGDV 360 DPVRWPNSYW RSLKVEWDES MASGRHERAS LWEIEPFNSV ATLPPPSLGQ NPSQKRRPVR 420 PTLVTCRNTP ESFHSTKLAR VLVHDQGGLK LREDEEGGTS WSLSGRAVKC EEVLKTPLMG 480 QDRTGAEGWL PMQQPEICPG VEIPHGLPRA DFTGVLTREP SQHNVEREKE HLRLCVQNYL 540 REPKEHKDSP SPRLSSTGYG KPWYGSEDMQ ISVNSSSIWD PTMKLPWPSN LPHVPSVPES 600 EHPSSPAPIW PATSAFPDAA ASLSRSLGAR GEFFPSWEDQ EGKVLAPTPP PSKPQCKLFG 660 FALNGDNAPN PAPSADLPKE AEMDVEPLSS GPTNSGGSNS QELAVKSGAV MKLPSFPVRS 720 FIKVHKQGEP GRSIDLTRYE SYTELRRALE QMFSLHGQLE TPAKQWQLVY TDHEGDVLLV 780 GDDPWGEFCS CVRSLKILSP TIEGGSEDEV QADLQDTTSP LMPGTK* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldv_A | 1e-127 | 54 | 401 | 9 | 359 | Auxin response factor 1 |
| 4ldw_A | 1e-127 | 54 | 401 | 9 | 359 | Auxin response factor 1 |
| 4ldw_B | 1e-127 | 54 | 401 | 9 | 359 | Auxin response factor 1 |
| 4ldx_A | 1e-127 | 54 | 401 | 9 | 359 | Auxin response factor 1 |
| 4ldx_B | 1e-127 | 54 | 401 | 9 | 359 | Auxin response factor 1 |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00574 | DAP | Transfer from AT5G62000 | Download |
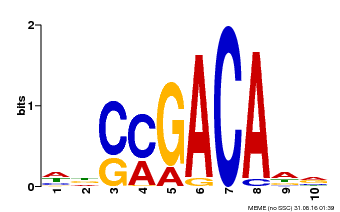
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024398385.1 | 0.0 | auxin response factor 7-like isoform X2 | ||||
| Refseq | XP_024398386.1 | 0.0 | auxin response factor 7-like isoform X2 | ||||
| Refseq | XP_024398387.1 | 0.0 | auxin response factor 7-like isoform X2 | ||||
| Refseq | XP_024398388.1 | 0.0 | auxin response factor 7-like isoform X2 | ||||
| TrEMBL | A0A2K1J7D0 | 0.0 | A0A2K1J7D0_PHYPA; Auxin response factor | ||||
| STRING | PP1S341_4V6.1 | 0.0 | (Physcomitrella patens) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G37020.2 | 1e-131 | auxin response factor 8 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Sphfalx0095s0033.1.p |



