 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sphfalx0119s0060.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Bryophyta; Sphagnophytina; Sphagnopsida; Sphagnales; Sphagnaceae; Sphagnum
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 541aa MW: 58625.8 Da PI: 6.1133 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 36.9 | 8.6e-12 | 14 | 61 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+ Ed +l ++k +G g+W+t ++ g+ R++k+ ++rw +yl
Sphfalx0119s0060.1.p 14 KGPWTEAEDRILSAYIKAHGAGCWRTLPKAAGLARCGKSSRLRWINYL 61
79********************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 49.2 | 1.2e-15 | 67 | 112 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg+ +++Ed+l++ ++ +lG++ W++Ia +++ gRt++++k++w+++l
Sphfalx0119s0060.1.p 67 RGNISPDEDDLIITLHSLLGNR-WSLIAGRIP-GRTDNEIKNYWNTHL 112
7899******************.*********.************996 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 1.1E-20 | 5 | 64 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 13.969 | 9 | 61 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.33E-25 | 11 | 108 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.4E-8 | 13 | 63 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.1E-9 | 14 | 61 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 3.63E-7 | 16 | 61 | No hit | No description |
| PROSITE profile | PS51294 | 26.326 | 62 | 116 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.5E-25 | 65 | 117 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 6.3E-16 | 66 | 114 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.3E-14 | 67 | 112 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.23E-11 | 71 | 112 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0010224 | Biological Process | response to UV-B | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:1900384 | Biological Process | regulation of flavonol biosynthetic process | ||||
| GO:1903086 | Biological Process | negative regulation of sinapate ester biosynthetic process | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 541 aa Download sequence Send to blast |
MGRAPCCDKQ GINKGPWTEA EDRILSAYIK AHGAGCWRTL PKAAGLARCG KSSRLRWINY 60 LRPDLKRGNI SPDEDDLIIT LHSLLGNRWS LIAGRIPGRT DNEIKNYWNT HLKRKLKSMG 120 LDPSGKHKLP SFPFFSTSST TSLAQENSSS HVNQAPKDES SLEPEEETPH VLHQNNSSPS 180 SLTSPPQFHD IKEIKKDRLS IMMRTAQVMA THSTMRTATH NSTEADQSKA LSFGNFSSEE 240 DSDRSSSLIP DPGQAAVAAR SSWEPTLENI VKAAQGQAPL YPSSPDSVLS TCKGLHKDLN 300 HHKSQASLVA PGHSIPQMVV AGATSMEITQ VPLHHHLPVR SNSNPLGIMV SEYNMPSTCS 360 TDFSTDASSL AEQNYSSSWN NSTCIELVHS ESSSGFYRND ASFNCNDSDS GYFNKSISGE 420 SLRGGAAAAT TVLDSAAAAA PADQDPYYTL MANLSSLKYT NTDQLSSISS SGSSHCGGYS 480 SPLENLFSPF PDYTPDSDDL QHEMQASLLG DHAWGGNLQP SHSSMDFLSN DDSLWEPCIM 540 * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 1e-24 | 14 | 118 | 7 | 110 | B-MYB |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 111 | 117 | LKRKLKS |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00479 | DAP | Transfer from AT4G38620 | Download |
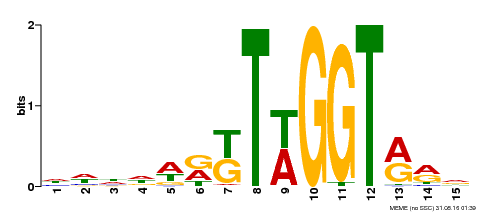
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G16720.1 | 2e-67 | myb domain protein 7 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Sphfalx0119s0060.1.p |



