 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sphfalx0144s0014.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Bryophyta; Sphagnophytina; Sphagnopsida; Sphagnales; Sphagnaceae; Sphagnum
|
||||||||
| Family | ZF-HD | ||||||||
| Protein Properties | Length: 272aa MW: 29726.7 Da PI: 9.3127 | ||||||||
| Description | ZF-HD family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | ZF-HD_dimer | 106.6 | 1.5e-33 | 75 | 131 | 3 | 60 |
ZF-HD_dimer 3 kvrYkeClkNhAaslGghavDGCgEfmpsegeegtaaalkCaACgCHRnFHRreveee 60
++rY+eC+kNhAaslG+havDGCgEfmp geeg+++alkCaACgCHRnFHRreve e
Sphfalx0144s0014.1.p 75 TTRYRECQKNHAASLGSHAVDGCGEFMPL-GEEGSPDALKCAACGCHRNFHRREVEGE 131
789*************************9.999*********************9865 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| ProDom | PD125774 | 3.0E-23 | 71 | 140 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| Pfam | PF04770 | 2.0E-30 | 76 | 128 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| TIGRFAMs | TIGR01566 | 2.2E-30 | 77 | 128 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| PROSITE profile | PS51523 | 26.856 | 78 | 127 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| SuperFamily | SSF46689 | 3.78E-20 | 195 | 269 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 2.8E-30 | 197 | 270 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01565 | 3.1E-25 | 208 | 264 | IPR006455 | Homeodomain, ZF-HD class |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 272 aa Download sequence Send to blast |
MRLLLTTSPQ LLHQPPLFHD STKLKMLKSG SNNNNNNNNG TAMDDSIQAL EITSPVNRTL 60 NSGNKDKDKK VAATTTRYRE CQKNHAASLG SHAVDGCGEF MPLGEEGSPD ALKCAACGCH 120 RNFHRREVEG EIATCDCHKK DAKKRGIQQV LAAATAMAYP SSVTQFQQGH KPPMTPIPMM 180 AMSTGAADSD EQEGGGGVGG GGSPSNIKKR FRTKFSTEQK DEMYNFAEKL GWKMQKHDEL 240 AVQEFCANLG VKRHVLKVWM HNNKHTMAKK T* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wh7_A | 4e-24 | 209 | 266 | 18 | 75 | ZF-HD homeobox family protein |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00258 | DAP | Transfer from AT2G02540 | Download |
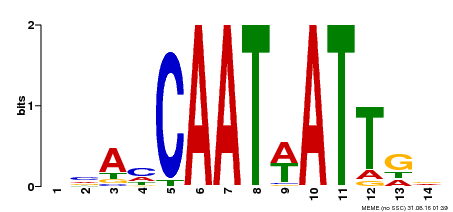
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024402412.1 | 1e-85 | zinc-finger homeodomain protein 1-like | ||||
| TrEMBL | A0A2K1J0V6 | 2e-84 | A0A2K1J0V6_PHYPA; Uncharacterized protein | ||||
| TrEMBL | A9RDY7 | 8e-85 | A9RDY7_PHYPA; Predicted protein | ||||
| STRING | PP1S3_539V6.1 | 4e-85 | (Physcomitrella patens) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G24660.1 | 1e-56 | homeobox protein 22 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Sphfalx0144s0014.1.p |



