 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sphfalx0148s0016.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Bryophyta; Sphagnophytina; Sphagnopsida; Sphagnales; Sphagnaceae; Sphagnum
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 662aa MW: 70863.3 Da PI: 4.5586 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 64.4 | 2.3e-20 | 260 | 309 | 2 | 55 |
AP2 2 gykGVrwdkkrgrWvAeIrd.psengkr.krfslgkfgtaeeAakaaiaarkkleg 55
+++GVr+++ +g+W+AeIrd + + r +lg+++taeeAa+a+++a+++++g
Sphfalx0148s0016.1.p 260 KFRGVRQRP-WGKWAAEIRDpF-----KgVRLWLGTYDTAEEAAQAYDKAARQIRG 309
89*******.**********55.....45************************998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 1.52E-33 | 260 | 317 | No hit | No description |
| SuperFamily | SSF54171 | 3.14E-22 | 260 | 317 | IPR016177 | DNA-binding domain |
| PROSITE profile | PS51032 | 23.881 | 260 | 317 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 8.2E-37 | 260 | 323 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 2.3E-31 | 260 | 317 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 2.2E-13 | 260 | 309 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 2.8E-10 | 261 | 272 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 2.8E-10 | 283 | 299 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0042991 | Biological Process | transcription factor import into nucleus | ||||
| GO:0048364 | Biological Process | root development | ||||
| GO:0048825 | Biological Process | cotyledon development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 662 aa Download sequence Send to blast |
MPGPKTLPKF PPAAMAAAPA VANKVKNKLC STKVLKRDLV FSRESELEDA ESWDHGKLQI 60 CPKRVRVICT DPDATDSSSD EEGNFRRSEV LIRNNSHRKR RQYVQQIDIR LAHQQQQGAA 120 FFSSCSASDA DSEEEATAAA VVDEDVPSYH SIFTAQAMMM RCSHNYASPI GGTARSSVVP 180 LLQQQQQQNK VFKSCGKAKK KASKIPKMPV LKKLQLDAAA TTSASSKPSA ASAQATAKSL 240 AAAAKTSSRG GGGEDGKINK FRGVRQRPWG KWAAEIRDPF KGVRLWLGTY DTAEEAAQAY 300 DKAARQIRGP QAHTNFPGFE ELELPTAANP DSSSGLKKRA AASPENPNGC TGVSNKKDVA 360 AAGKCVAAVA FNSEKKPLCP AVAEIEFLTD RVSSSEEEEK SCHTAVFEPE VNDDGGSPSA 420 SERSEGSALT RSMEELGEDD DDVHHPNLRE VVGQGQSLPC YDGFPHPDEE CANFLMCSPS 480 SALDGCSTSF SELSSGSIAD NSDAFSCDPF TTSGRSMGVE EESHGLAEST NGNTSHTPNI 540 VDQTSLGVNL AVVQKDNAQE KESEDKDTSL DSELVGKEAR GGEDKDAELQ NLADLSQVFF 600 SDDEFLFDIP DCDGDDGGLI MMEFEAGFDL LCDGEKIMDL GLDGGNEAMN WFSAPDNIAI 660 A* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gcc_A | 2e-20 | 261 | 317 | 3 | 60 | ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 1 |
| 5wx9_A | 9e-20 | 249 | 317 | 6 | 72 | Ethylene-responsive transcription factor ERF096 |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00027 | PBM | Transfer from AT4G23750 | Download |
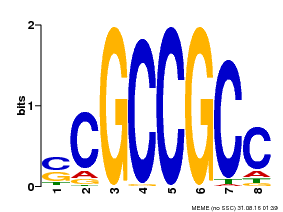
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G23750.2 | 8e-20 | cytokinin response factor 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Sphfalx0148s0016.1.p |



