 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sphfalx0194s0025.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Bryophyta; Sphagnophytina; Sphagnopsida; Sphagnales; Sphagnaceae; Sphagnum
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 968aa MW: 108025 Da PI: 4.8147 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 43.1 | 9.7e-14 | 8 | 53 | 2 | 48 |
SSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 2 grWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
g W Ede+l+ av ++G+++W++I++ + ++++kqck rw+ +l
Sphfalx0194s0025.1.p 8 GVWKNTEDEILKAAVMKYGKNQWARISSLLV-RKSAKQCKARWYEWL 53
78*****************************.************986 PP
| |||||||
| 2 | Myb_DNA-binding | 32.9 | 1.5e-10 | 60 | 103 | 2 | 48 |
SSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 2 grWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
WT eEde+l+++ k++++ W+tIa +g Rt+ qc +r+ k+l
Sphfalx0194s0025.1.p 60 TEWTREEDEKLLHLAKLMPTQ-WRTIAPIVG--RTPAQCLERYEKLL 103
68*****************99.********8..**********9986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 22.008 | 2 | 57 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 5.6E-18 | 5 | 56 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 7.0E-15 | 6 | 55 | IPR001005 | SANT/Myb domain |
| Pfam | PF13921 | 4.5E-13 | 10 | 70 | No hit | No description |
| CDD | cd00167 | 5.77E-12 | 10 | 53 | No hit | No description |
| SuperFamily | SSF46689 | 6.11E-21 | 33 | 108 | IPR009057 | Homeodomain-like |
| CDD | cd11659 | 9.06E-31 | 55 | 106 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 3.9E-14 | 57 | 104 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.5E-12 | 58 | 105 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS51294 | 16.529 | 58 | 107 | IPR017930 | Myb domain |
| Pfam | PF11831 | 4.8E-56 | 407 | 663 | IPR021786 | Pre-mRNA splicing factor component Cdc5p/Cef1 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009870 | Biological Process | defense response signaling pathway, resistance gene-dependent | ||||
| GO:0010204 | Biological Process | defense response signaling pathway, resistance gene-independent | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0009507 | Cellular Component | chloroplast | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 968 aa Download sequence Send to blast |
MRIMIKGGVW KNTEDEILKA AVMKYGKNQW ARISSLLVRK SAKQCKARWY EWLDPSIKKT 60 EWTREEDEKL LHLAKLMPTQ WRTIAPIVGR TPAQCLERYE KLLDAACAKD ENYEPADDPR 120 KLRPGEIDPN PESKPARPDP VDMDEDEKEM LSEARARLAN TRGKKAKRKA REKQLEEARR 180 LASLQKRREL KAAGIDGRHR RRKQKGIDYN AEIPFEKKPP PGFFEVPHEA PVEQPQFPTT 240 IEELEGKRRV DIESQLRKQD VARNKIAERR DAPAAVMQVN KLNDPEAVRK RSKLMLPEPQ 300 ISDRELEEIA KMGYASDLIS GEDGLVEGSG VTRSLLSNYN QTPARPGMTP LRTPQRTPGG 360 KGDAIMMEAE NLARLRETQT PLFGGENPDL HPSDFSGVTP KKKEIQTPNP IATPLRTPSG 420 ATGATPRLSM TPAKDVAVYG MTPKGTPIRD ELHINEGMDT PSLESSKAEK LRQAELRRHL 480 RMGLGDLPAP KNEYQIMVPD LPEEEPVEGD DMEADMSDVL ARQHAEEAAK QAALLRKRSK 540 VLQRVLPRPP PAAIEMMRAS LAYGEDGKAA PAPVTLIDHA DLLVRAELVA LLEYDAAKYP 600 MLEEESSKAK KKGAGKNAAN GNPQVPVPVL EDFDEADLND AGLLIEEELG YIRSAMGHED 660 ATLDDYAEAR DACVDDLMYL PTRDSFGLAS VASTSDRLAA LQYQFENVKK QMEAETRKAV 720 RLEQKIKVLT QGYQIRADAL WRQIELTFKE ADTLGTELEC FWALHRQEQL AAPRRIENLQ 780 DAVKQQSEKE RMLQLRYENL LVEQENLRGF IHDHDAAAAK AAEEKEIAEK KAAEEKQAME 840 DMNTAEEARA AEEADAAKKR EEEDKPMADQ SDDKSVEEQS AVEKALQEKD NTDAPTLATA 900 VVEAEEAKAN DMVEASKAEA LSLEGPTETD KEGPTAMEGL EDEPVGPPPP PPESQDDGDD 960 TSSDRRS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5mqf_L | 0.0 | 2 | 809 | 3 | 800 | Cell division cycle 5-like protein |
| 5xjc_L | 0.0 | 2 | 809 | 3 | 800 | Cell division cycle 5-like protein |
| 5yzg_L | 0.0 | 2 | 809 | 3 | 800 | Cell division cycle 5-like protein |
| 5z56_L | 0.0 | 2 | 809 | 3 | 800 | Cell division cycle 5-like protein |
| 5z57_L | 0.0 | 2 | 809 | 3 | 800 | Cell division cycle 5-like protein |
| 5z58_L | 0.0 | 2 | 809 | 3 | 800 | Cell division cycle 5-like protein |
| 6ff4_L | 0.0 | 2 | 809 | 3 | 800 | Cell division cycle 5-like protein |
| 6ff7_L | 0.0 | 2 | 809 | 3 | 800 | Cell division cycle 5-like protein |
| 6icz_L | 0.0 | 2 | 809 | 3 | 800 | Cell division cycle 5-like protein |
| 6id0_L | 0.0 | 2 | 809 | 3 | 800 | Cell division cycle 5-like protein |
| 6id1_L | 0.0 | 2 | 809 | 3 | 800 | Cell division cycle 5-like protein |
| 6qdv_O | 0.0 | 2 | 809 | 3 | 800 | Cell division cycle 5-like protein |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 185 | 202 | KRRELKAAGIDGRHRRRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
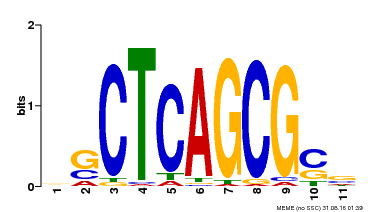
| UniProt | Component of the MAC complex that probably regulates defense responses through transcriptional control and thereby is essential for plant innate immunity. Possesses a sequence specific DNA sequence 'CTCAGCG' binding activity. Involved in mRNA splicing and cell cycle control. May also play a role in the response to DNA damage. {ECO:0000250|UniProtKB:Q99459, ECO:0000269|PubMed:17298883, ECO:0000269|PubMed:17575050, ECO:0000269|PubMed:8917598}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00028 | SELEX | Transfer from AT1G09770 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024370979.1 | 0.0 | cell division cycle 5-like protein | ||||
| Swissprot | P92948 | 0.0 | CDC5L_ARATH; Cell division cycle 5-like protein | ||||
| TrEMBL | A0A176WFB4 | 0.0 | A0A176WFB4_MARPO; Uncharacterized protein | ||||
| STRING | PP1S641_1V6.1 | 0.0 | (Physcomitrella patens) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09770.1 | 0.0 | cell division cycle 5 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Sphfalx0194s0025.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



