 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sphfalx0217s0041.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Bryophyta; Sphagnophytina; Sphagnopsida; Sphagnales; Sphagnaceae; Sphagnum
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 571aa MW: 61073.8 Da PI: 7.4119 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 57.6 | 2.9e-18 | 23 | 68 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+g+W++eEd l ++v+++G ++W++I++ ++ gR++k+c++rw +
Sphfalx0217s0041.1.p 23 KGPWSPEEDAALQQLVEKHGARNWSLISKGIP-GRSGKSCRLRWCNQ 68
79******************************.***********985 PP
| |||||||
| 2 | Myb_DNA-binding | 49.6 | 9.1e-16 | 77 | 119 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
++T+ Ed +++a++++G++ W+tIar ++ gRt++ +k++w++
Sphfalx0217s0041.1.p 77 PFTAFEDSAIIQAHHLHGNK-WATIARILP-GRTDNAIKNHWNST 119
79999***************.*********.***********986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 26.193 | 18 | 73 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 3.75E-31 | 20 | 116 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 5.0E-17 | 22 | 71 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.8E-18 | 23 | 68 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.3E-24 | 24 | 76 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.29E-15 | 25 | 67 | No hit | No description |
| PROSITE profile | PS51294 | 19.696 | 74 | 124 | IPR017930 | Myb domain |
| SMART | SM00717 | 4.1E-13 | 74 | 122 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 4.0E-13 | 77 | 119 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 4.5E-21 | 77 | 123 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.00E-10 | 77 | 120 | No hit | No description |
| PROSITE profile | PS50873 | 8.614 | 377 | 570 | IPR002016 | Haem peroxidase, plant/fungal/bacterial |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006979 | Biological Process | response to oxidative stress | ||||
| GO:0055114 | Biological Process | oxidation-reduction process | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0004601 | Molecular Function | peroxidase activity | ||||
| GO:0020037 | Molecular Function | heme binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 571 aa Download sequence Send to blast |
MALVSSATKA SPTQVLEEFD RIKGPWSPEE DAALQQLVEK HGARNWSLIS KGIPGRSGKS 60 CRLRWCNQLS PSVQHRPFTA FEDSAIIQAH HLHGNKWATI ARILPGRTDN AIKNHWNSTL 120 RRRHLAETTT SSERERRSLR RGAAETTTAD DGVSSLIRIS NNNNDEEEIG SFDGRKRNSN 180 EISISISTDG SLQEESGWEV DSHRLKKLSF GKTDNNDDHH AQQQQHRSNS KRLSISGADD 240 DAAAPQRLRK LSFAGSDSSA MSSFLPPSDH DIPAKLNKKH GFGSESSTLL MPAPPSLFRP 300 VPRASAFNTF TPTPAPPPPA PPLSSYCHAS SSALAGVQQD SIVERHKDMI HNQAEEEALA 360 TVDPTTSLSL SPPGTDQSCT TSQVSAAAEQ QVVEKKQPPT TPMSSSAFTE SSTAAHVYQA 420 PSMNGLFFGP NSETCANLLS VAVKSAVAQV LAPVFPPPQP VAQGGMGANW ASSSSSSSAL 480 APFGSSINSS SSYGLDAAVN AGLLAMMRDM VAKEVHNYMA AVHSSSCFPS FHAFATHHEH 540 ATSLASRATT FSNQPEFLGA MATTAPRKAV * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 8e-37 | 20 | 123 | 4 | 107 | B-MYB |
| 1mse_C | 8e-37 | 22 | 123 | 3 | 104 | C-Myb DNA-Binding Domain |
| 1msf_C | 8e-37 | 22 | 123 | 3 | 104 | C-Myb DNA-Binding Domain |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00632 | PBM | Transfer from PK28565.1 | Download |
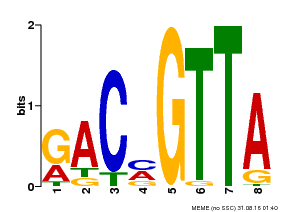
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G23290.1 | 6e-56 | myb domain protein 70 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Sphfalx0217s0041.1.p |



