 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sphfalx0244s0009.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Bryophyta; Sphagnophytina; Sphagnopsida; Sphagnales; Sphagnaceae; Sphagnum
|
||||||||
| Family | GATA | ||||||||
| Protein Properties | Length: 360aa MW: 39527.8 Da PI: 4.8007 | ||||||||
| Description | GATA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GATA | 54.7 | 1.4e-17 | 287 | 321 | 1 | 35 |
GATA 1 CsnCgttkTplWRrgpdgnktLCnaCGlyyrkkgl 35
C++C+t Tp+WR gp g+ktLCnaCG++y++ +l
Sphfalx0244s0009.1.p 287 CTHCQTQRTPQWRTGPMGPKTLCNACGVRYKSGRL 321
*******************************9986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00401 | 7.6E-18 | 281 | 335 | IPR000679 | Zinc finger, GATA-type |
| PROSITE profile | PS50114 | 11.767 | 281 | 317 | IPR000679 | Zinc finger, GATA-type |
| SuperFamily | SSF57716 | 5.23E-15 | 285 | 344 | No hit | No description |
| CDD | cd00202 | 2.61E-14 | 286 | 335 | No hit | No description |
| Gene3D | G3DSA:3.30.50.10 | 1.2E-14 | 286 | 319 | IPR013088 | Zinc finger, NHR/GATA-type |
| PROSITE pattern | PS00344 | 0 | 287 | 312 | IPR000679 | Zinc finger, GATA-type |
| Pfam | PF00320 | 2.4E-15 | 287 | 321 | IPR000679 | Zinc finger, GATA-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0009416 | Biological Process | response to light stimulus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 360 aa Download sequence Send to blast |
MDSDALLLPA GLVAFDNSLK KQQQQQKPLT VAAADAAGSS FADIFQIDDL LDFSNEDIAG 60 PIGEAVHVLK PETIGGCLTP FLENGTELQH LAPKAHKLEG EEEEAAIEAD LCLPQYDDLE 120 ELEWASRFLE DSFPVTGDDE SHKTWSDDNG SAISIGEKET ELSALFMKKQ HQYYEKAAAS 180 PVSVLEHQAS ATSSELTGSD HSLDFCVPGR ARSKRSRSEG KKWTSGILSS TTTSCGTNRV 240 SASPSQWSAG EEEDTVEEGE EWQSCNFQKA KQSRKSSSQQ DGEVLRCTHC QTQRTPQWRT 300 GPMGPKTLCN ACGVRYKSGR LLPEYRPASS PAYIPHKHSN SHKKILEMRQ QREMMEASM* |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00048 | PBM | Transfer from AT4G32890 | Download |
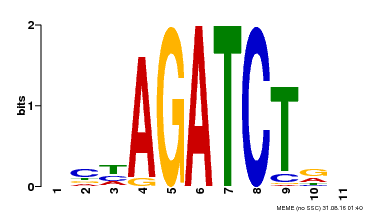
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G32890.1 | 1e-45 | GATA transcription factor 9 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Sphfalx0244s0009.1.p |



