 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sphfalx0246s0020.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Bryophyta; Sphagnophytina; Sphagnopsida; Sphagnales; Sphagnaceae; Sphagnum
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 573aa MW: 61638.3 Da PI: 6.7872 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 49.9 | 5.6e-16 | 455 | 501 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L+el+P++ K +Ka++L +A+eY++ Lq
Sphfalx0246s0020.1.p 455 VHNLSERRRRDRINEKMKALQELIPNS-----NKTDKASMLDEAIEYLRTLQ 501
6*************************7.....5******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.280.10 | 8.2E-19 | 449 | 510 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 3.27E-19 | 449 | 510 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 18.19 | 451 | 500 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 1.81E-17 | 454 | 505 | No hit | No description |
| Pfam | PF00010 | 1.6E-13 | 455 | 501 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.5E-17 | 457 | 506 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0031539 | Biological Process | positive regulation of anthocyanin metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 573 aa Download sequence Send to blast |
MQCEGSNGCK LQWQPSYGSS GDKGQVADDG TLEAVVNETA AETTLTPKIN YHDHETVSWL 60 QYPLDDTLER KYCSDFFGEL PSSNMQVVKG SFVGQGAGCL QCMPIPNNMV TDAVVNKATS 120 TEAAMILHAG HAAGVIVQTG EEVFNKVQTL QPLVSRWQPY SLTPNSSKGF GVYPTVSQAP 180 LRTSSPASRP PLAPMAPPKP QSRGGMDNMH ADSARKLASL NSLHSSRSAV TMKNHQSVGV 240 LTGTPSSMFS KQMSVKVNAE AGTLTSSSIA ESATTGRSGM GCQNNIEIHL QGSDSQQQSQ 300 GVDARGHCPV IRKEVDTLVL EGEAMLPVPV PSGRKEVDMP VLEQETVLGT TKPTEQDTLQ 360 KSPVTASAAL ESADKWMSQG RQDPDAQELT ITSSLGGCGN SAERAKQAST SNKRKAVVTE 420 EPECQSEDGD DESADSKRRP EGRASTTKRT RAAEVHNLSE RRRRDRINEK MKALQELIPN 480 SNKTDKASML DEAIEYLRTL QLQIQMISMR TGLRMPPMVR SAGTQYLQIP SLEMTSIPSL 540 GLGMGRGLGM SRMDMAASTS NLQPHGYLKH SS* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 459 | 464 | ERRRRD |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00081 | ChIP-seq | Transfer from AT1G09530 | Download |
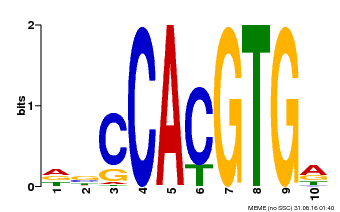
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09530.2 | 1e-39 | phytochrome interacting factor 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Sphfalx0246s0020.1.p |



