 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Spipo0G0019400 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Alismatales; Araceae; Lemnoideae; Spirodela
|
||||||||
| Family | ZF-HD | ||||||||
| Protein Properties | Length: 221aa MW: 23826.5 Da PI: 7.36 | ||||||||
| Description | ZF-HD family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | ZF-HD_dimer | 104.8 | 5.4e-33 | 19 | 73 | 3 | 58 |
ZF-HD_dimer 3 kvrYkeClkNhAaslGghavDGCgEfmpsegeegtaaalkCaACgCHRnFHRreve 58
+vrY+eClkNhAa++Ggh++DGCgEfmps geegt++alkCaAC CHR+FHR+eve
Spipo0G0019400 19 TVRYRECLKNHAARIGGHVLDGCGEFMPS-GEEGTTEALKCAACACHRSFHRKEVE 73
79**************************9.999********************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| ProDom | PD125774 | 3.0E-21 | 18 | 70 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| Pfam | PF04770 | 1.3E-30 | 20 | 72 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| TIGRFAMs | TIGR01566 | 4.7E-30 | 21 | 72 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| PROSITE profile | PS51523 | 27.401 | 22 | 71 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| SuperFamily | SSF46689 | 8.97E-19 | 148 | 215 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 1.3E-30 | 149 | 219 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01565 | 2.9E-27 | 158 | 215 | IPR006455 | Homeodomain, ZF-HD class |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 221 aa Download sequence Send to blast |
GTGPSAASSV PRTLGSGPTV RYRECLKNHA ARIGGHVLDG CGEFMPSGEE GTTEALKCAA 60 CACHRSFHRK EVEGGEEAEG LFAGGHDDEE NNYSGGTTTT ESSSEEMQAH MLAAAGVTPF 120 RQVAHSSHPF QASHAVGMGE MQHYQIAGGP GSASGSAKKR HRTKFTAEQK EMMAEFAERV 180 GWRMQKQDDG ALERFCREAG VSRQVFKVWM HNNKNVVLKS K |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wh5_A | 4e-25 | 158 | 214 | 17 | 73 | ZF-HD homeobox family protein |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00268 | DAP | Transfer from AT2G18350 | Download |
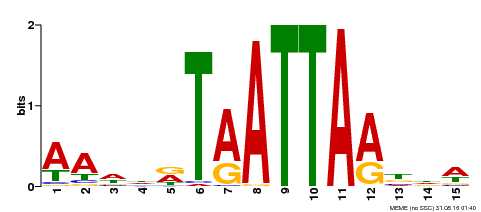
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| TrEMBL | A0A200R7P2 | 3e-59 | A0A200R7P2_9MAGN; Homeobox domain | ||||
| STRING | XP_010539899.1 | 2e-59 | (Tarenaya hassleriana) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP815 | 35 | 145 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G18350.1 | 5e-50 | homeobox protein 24 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Spipo0G0019400 |



