 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Spipo0G0095000 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Alismatales; Araceae; Lemnoideae; Spirodela
|
||||||||
| Family | HSF | ||||||||
| Protein Properties | Length: 257aa MW: 28568.9 Da PI: 7.4518 | ||||||||
| Description | HSF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HSF_DNA-bind | 114.1 | 9.2e-36 | 42 | 134 | 2 | 103 |
HHHHHHHHHCTGGGTTTSEESSSSSEEEES-HHHHHHHTHHHHSTT--HHHHHHHHHHTTEEE---SSBTTTTXTTSEEEEESXXXXXXXXXXXX CS
HSF_DNA-bind 2 FlkklyeiledeelkeliswsengnsfvvldeeefakkvLpkyFkhsnfaSFvRQLnmYgFkkvkdeekkskskekiweFkhksFkkgkkellek 96
Fl+k+y++++d+++++++sw ++ ++fvv+ + efa+++Lp+yFkh+nf+SFvRQLn+YgF+k+ e+ weF+++ F+kg+k+ll++
Spipo0G0095000 42 FLTKTYQLVDDPATDHVVSWGDDHSTFVVWRPPEFARDILPNYFKHNNFSSFVRQLNTYGFRKIVPER---------WEFANELFRKGEKHLLCD 127
9****************************************************************999.........****************** PP
XXXXXXX CS
HSF_DNA-bind 97 ikrkkse 103
i+r+k++
Spipo0G0095000 128 IHRRKTS 134
****986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.10 | 9.2E-38 | 34 | 126 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SMART | SM00415 | 5.2E-57 | 38 | 131 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| SuperFamily | SSF46785 | 6.8E-34 | 39 | 131 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| PRINTS | PR00056 | 8.8E-20 | 42 | 65 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| Pfam | PF00447 | 2.4E-31 | 42 | 131 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 8.8E-20 | 80 | 92 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PROSITE pattern | PS00434 | 0 | 81 | 105 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 8.8E-20 | 93 | 105 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0008356 | Biological Process | asymmetric cell division | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 257 aa Download sequence Send to blast |
MALAAAPVAS PLEPCSGSEG GDCGGAATLL TLEPHRAVPA PFLTKTYQLV DDPATDHVVS 60 WGDDHSTFVV WRPPEFARDI LPNYFKHNNF SSFVRQLNTY GFRKIVPERW EFANELFRKG 120 EKHLLCDIHR RKTSSSPSTS PHAPLHLPFQ HYHPHTPGAN ASTAASYFSS FSTSPDEHIS 180 PSGGWCDSFS SPISSPRVAP PGSGLLEDNE RLRRANATLL SELAHMRKLY NDIIYFVQNH 240 PRETFNGERE RGKKVY* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2ldu_A | 4e-24 | 38 | 136 | 17 | 125 | Heat shock factor protein 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional regulator that specifically binds DNA sequence 5'-AGAAnnTTCT-3' known as heat shock promoter elements (HSE). | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00187 | DAP | Transfer from AT1G46264 | Download |
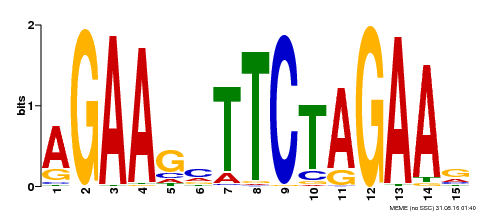
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK356002 | 3e-95 | AK356002.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv1028L19. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011017186.1 | 3e-92 | PREDICTED: heat stress transcription factor B-4-like | ||||
| Refseq | XP_012466618.1 | 2e-92 | PREDICTED: heat stress transcription factor B-4-like isoform X1 | ||||
| Refseq | XP_016706734.1 | 2e-92 | PREDICTED: heat stress transcription factor B-4-like isoform X1 | ||||
| Swissprot | Q9C635 | 2e-89 | HSFB4_ARATH; Heat stress transcription factor B-4 | ||||
| TrEMBL | A0A2P5SGA1 | 4e-91 | A0A2P5SGA1_GOSBA; Uncharacterized protein | ||||
| STRING | Gorai.002G135200.1 | 1e-91 | (Gossypium raimondii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP121 | 37 | 394 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G46264.1 | 7e-88 | heat shock transcription factor B4 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Spipo0G0095000 |



