 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Spipo14G0026000 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Alismatales; Araceae; Lemnoideae; Spirodela
|
||||||||
| Family | Nin-like | ||||||||
| Protein Properties | Length: 887aa MW: 97371.4 Da PI: 6.3082 | ||||||||
| Description | Nin-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | RWP-RK | 89.5 | 2.6e-28 | 579 | 630 | 1 | 52 |
RWP-RK 1 aekeisledlskyFslpikdAAkeLgvclTvLKriCRqyGIkRWPhRkiksl 52
aek+isle+l+ yF++++kdAAk+Lgvc+T++KriCRq+GI+RWP+Rkik++
Spipo14G0026000 579 AEKTISLEVLKCYFAGSLKDAAKSLGVCPTTMKRICRQHGISRWPSRKIKKV 630
589***********************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51519 | 17.177 | 567 | 650 | IPR003035 | RWP-RK domain |
| Pfam | PF02042 | 1.1E-24 | 582 | 630 | IPR003035 | RWP-RK domain |
| SuperFamily | SSF54277 | 1.24E-18 | 793 | 876 | No hit | No description |
| SMART | SM00666 | 1.6E-18 | 794 | 873 | IPR000270 | PB1 domain |
| PROSITE profile | PS51745 | 18.807 | 794 | 873 | IPR000270 | PB1 domain |
| Gene3D | G3DSA:3.10.20.240 | 2.0E-22 | 794 | 872 | No hit | No description |
| Pfam | PF00564 | 1.6E-13 | 795 | 872 | IPR000270 | PB1 domain |
| CDD | cd06407 | 7.05E-32 | 795 | 872 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0010118 | Biological Process | stomatal movement | ||||
| GO:0010167 | Biological Process | response to nitrate | ||||
| GO:0042128 | Biological Process | nitrate assimilation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 887 aa Download sequence Send to blast |
MSEGGEEKPD DGPPLPLPPP VISGDAMDVD LELESSWSFD AGDFLRDSSI EYFLLSSSPL 60 GPLQSSLQFP QASPPQSPLL SFEQRVTEVS DAVPEISNLV PGICDPDDDV LPRSSSKARH 120 ISKAAKEETY DVSRIIKEKM THALRCFKDS IDQHVLVQVW APVKNGDRRV LTTSEQPFVL 180 DPHSISLLQY RTISLTYLFS VDGHSDGDLG LPGRVFSQQF PEWTPDVQYY SSKEYPRLTH 240 ALNHNVKGSL ALPIFESFYE TCVGVVELIM TSPKINYASE VDKVCKALEA VNLKSSKILG 300 HPSVQILNEG HQTAMSEILE VFTTVCEMHK LPLAQTWVPC KHGRCLPHDG DKNIIALGHD 360 KSCLEQSCMS TTNAAFYIVE PRMWGFRDAC AEHRLQKGQG VVGMAYSLRR PCFSSNITQF 420 GKNSYPLVHY ARMFGLVGCL AVCLQSCHTG NDVYVLEFFL PSECRSEMQQ LALLETISTT 480 IRQLIRSLKV VFKTETQEDI FDEILHMNGD LEDAPICNEA HKPCEGASVE IKGPEALTAS 540 RTDKKNTQKS DLKKGMTPAM MESQNPASPK TQERRRGKAE KTISLEVLKC YFAGSLKDAA 600 KSLGVCPTTM KRICRQHGIS RWPSRKIKKV NRSLSKLKTV IESVQGTGGA FDLQNAARVW 660 PQQLHKSHQN KSLENGNLNE LVGIHEGNEP PGIENNTTKA ASRQGQPSEV NSPQCLTKGQ 720 PSEGTEARPP SQGSCPGNSA NRRSSKEVPP SSPNAAHGVN PMLVEDSGSS KDLKSLCLSL 780 DDACPDALLP SASSVKVKAN YRGDIIRFRV KSSDGVFLLK EQVAKRLKLD LGAFDLKYLD 840 DDHEWVLLAC DADLEECLEV NGSQAIRLSV TDVASNPESF SGSSGG* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00449 | DAP | Transfer from AT4G24020 | Download |
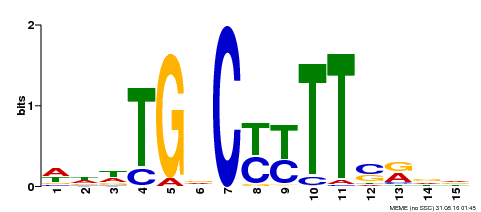
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008777047.1 | 0.0 | protein NLP3-like isoform X1 | ||||
| Swissprot | Q5NB82 | 0.0 | NLP3_ORYSJ; Protein NLP3 | ||||
| TrEMBL | A0A1D1XCV6 | 0.0 | A0A1D1XCV6_9ARAE; Protein NLP3 (Fragment) | ||||
| STRING | XP_008777047.1 | 0.0 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP4778 | 36 | 60 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G64530.1 | 0.0 | Nin-like family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Spipo14G0026000 |



