 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Spipo15G0009700 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Alismatales; Araceae; Lemnoideae; Spirodela
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 261aa MW: 27120.2 Da PI: 5.153 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 64.3 | 2.5e-20 | 75 | 123 | 3 | 55 |
AP2 3 ykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
++GVr+++ +g+WvAeIr p++ r+r +lg+f+taeeAa a+++a+ kl+g
Spipo15G0009700 75 FRGVRQRH-WGKWVAEIRLPKN---RTRLWLGTFDTAEEAALAYDRAAYKLRG 123
8******9.**********965...5*************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF54171 | 3.99E-23 | 74 | 132 | IPR016177 | DNA-binding domain |
| PROSITE profile | PS51032 | 22.695 | 74 | 131 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 2.5E-36 | 74 | 137 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 8.3E-32 | 74 | 132 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 6.6E-11 | 75 | 86 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 1.59E-31 | 75 | 133 | No hit | No description |
| Pfam | PF00847 | 3.9E-14 | 75 | 123 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 6.6E-11 | 97 | 113 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009611 | Biological Process | response to wounding | ||||
| GO:0009736 | Biological Process | cytokinin-activated signaling pathway | ||||
| GO:0009873 | Biological Process | ethylene-activated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0045595 | Biological Process | regulation of cell differentiation | ||||
| GO:0071472 | Biological Process | cellular response to salt stress | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 261 aa Download sequence Send to blast |
MLPPGECSFL SQLLSAGHGV DHLSAAQLRQ IQTHSAARQA SFLGPRGQPM KHPGAAAAIS 60 TPLVKSGGGG GARLFRGVRQ RHWGKWVAEI RLPKNRTRLW LGTFDTAEEA ALAYDRAAYK 120 LRGDFARLNF PELRGRCDSL SGDLASGGGG GSGSPLPSAV DAKLQAICES LAASNQPPAT 180 ARSGGGGEEE EEATAAPATA GDEAVGKQEA HLSLSPESGD SSPASEMETL DFSDPAWGES 240 ESMLLGKYPS LEIDWDAILS * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2gcc_A | 1e-19 | 75 | 131 | 6 | 63 | ATERF1 |
| 3gcc_A | 1e-19 | 75 | 131 | 6 | 63 | ATERF1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00026 | PBM | Transfer from AT1G78080 | Download |
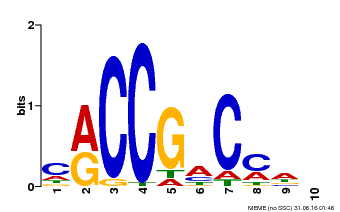
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_029123870.1 | 1e-65 | LOW QUALITY PROTEIN: ethylene-responsive transcription factor RAP2-13 | ||||
| TrEMBL | A0A3S3NIS5 | 1e-64 | A0A3S3NIS5_9MAGN; Ethylene-responsive transcription factor RAP2-4-like protein | ||||
| STRING | POPTR_0005s16690.1 | 9e-63 | (Populus trichocarpa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP382 | 35 | 203 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G39780.1 | 2e-38 | ERF family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Spipo15G0009700 |



