 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Spipo15G0027800 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Alismatales; Araceae; Lemnoideae; Spirodela
|
||||||||
| Family | GRAS | ||||||||
| Protein Properties | Length: 664aa MW: 70644.4 Da PI: 4.9998 | ||||||||
| Description | GRAS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GRAS | 426.8 | 1.7e-130 | 244 | 649 | 1 | 374 |
GRAS 1 lvelLlecAeavssgdlelaqalLarlselaspdgdpmqRlaayfteALaarlarsvselykalppsetseknsseelaalkl...fsevsPil 91
lv++L++cAeav+++++++a+al++++ l ++g +m+++a+yf+eALa+r++r ++p ++++ +++++ + l f+e +P+l
Spipo15G0027800 244 LVHTLMACAEAVQQDNFKAAEALVKQIGVLTMSQGGAMRKVATYFAEALARRIYR--------IRPGQDQS--LIDAAFEDILqmnFYESCPYL 327
689****************************************************........44444332..3333332222335******** PP
GRAS 92 kfshltaNqaIleavegeervHiiDfdisqGlQWpaLlqaLasRpegppslRiTgvgspesgskeeleetgerLakfAeelgvpfefnvlvakr 185
kf+h+taNqaIlea++g++rvH+iDf+++qG QWpaL+qaLa Rp+gpps+R+Tg+g+p+++++++l+++g++La++Ae+++v+fe++ +va +
Spipo15G0027800 328 KFAHFTANQAILEAFAGKSRVHVIDFSMKQGQQWPALMQALALRPGGPPSFRLTGIGPPQPDDSDALQQVGWKLAQLAESMHVEFEYRSFVAAS 421
********************************************************************************************** PP
GRAS 186 ledleleeLrvkp............................gEalaVnlvlqlhrlldesvsleserdevLklvkslsPkvvvvveqeadhnse 251
l+dle+ +L++ + +E+la+n+vl+lh++l++++++e+ vL +v+ l+Pk+v+vveqea+hn++
Spipo15G0027800 422 LADLEPYMLELASsarsssssggsrrkgdsagggssgeeqeEEVLAINSVLELHKMLARPGAVEK----VLGTVRALRPKIVTVVEQEANHNGP 511
*********554444555555555566666666666667779***********************....************************* PP
GRAS 252 sFlerflealeyysalfdsleak..............lpreseerikvErellgreivnvvacegaerrerhetlekWrerleeaGFkpvplse 331
F+erf eal+yys++fdsle ++e+ + +++ +++lg++i+nvvaceg er+erhet ++Wr r+e+aGF+pv++++
Spipo15G0027800 512 VFVERFNEALHYYSTMFDSLEGCaapppsaaglsgaaDAAEAGQIHVMTEVYLGQQICNVVACEGPERTERHETIGQWRGRMEAAGFEPVHIGS 605
********************8888999999998886433445889999********************************************** PP
GRAS 332 kaakqaklllrkvk.sdgyrveeesgslvlgWkdrpLvsvSaWr 374
+a kqa++ll+ ++ +dgyrvee++g+l+lgW++rpL+++SaWr
Spipo15G0027800 606 NAFKQASMLLAVFAgGDGYRVEEKDGCLMLGWHTRPLITTSAWR 649
*******************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF12041 | 6.9E-33 | 46 | 123 | IPR021914 | Transcriptional factor DELLA, N-terminal |
| SMART | SM01129 | 8.8E-31 | 46 | 129 | No hit | No description |
| PROSITE profile | PS50985 | 62.909 | 218 | 628 | IPR005202 | Transcription factor GRAS |
| Pfam | PF03514 | 5.9E-128 | 244 | 649 | IPR005202 | Transcription factor GRAS |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009867 | Biological Process | jasmonic acid mediated signaling pathway | ||||
| GO:0009938 | Biological Process | negative regulation of gibberellic acid mediated signaling pathway | ||||
| GO:0010187 | Biological Process | negative regulation of seed germination | ||||
| GO:0010218 | Biological Process | response to far red light | ||||
| GO:0042176 | Biological Process | regulation of protein catabolic process | ||||
| GO:0042538 | Biological Process | hyperosmotic salinity response | ||||
| GO:2000033 | Biological Process | regulation of seed dormancy process | ||||
| GO:2000377 | Biological Process | regulation of reactive oxygen species metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 664 aa Download sequence Send to blast |
MKREYTDGRD CGGGAGGDMA QQQQQGFSGK NKMAMMMAEQ QDAGVDEHLA VLGYKVRSSD 60 MADVAQKLEQ LEMAMVSGAV VGGGGAPDDN SILTHLSSET VHYNPSDLST WLETMLSELN 120 APPAPSPTAK APSSSSSSSS TAADNPLRAP PAGPSFSRPP ALLPDFPVPS IAGIDFPGSG 180 RPSSDSDLHA IPGGRVMYGN SDSADTEPRD RKRLRTSPAA AAPPSPESPV QVVLVDSQET 240 GIRLVHTLMA CAEAVQQDNF KAAEALVKQI GVLTMSQGGA MRKVATYFAE ALARRIYRIR 300 PGQDQSLIDA AFEDILQMNF YESCPYLKFA HFTANQAILE AFAGKSRVHV IDFSMKQGQQ 360 WPALMQALAL RPGGPPSFRL TGIGPPQPDD SDALQQVGWK LAQLAESMHV EFEYRSFVAA 420 SLADLEPYML ELASSARSSS SSGGSRRKGD SAGGGSSGEE QEEEVLAINS VLELHKMLAR 480 PGAVEKVLGT VRALRPKIVT VVEQEANHNG PVFVERFNEA LHYYSTMFDS LEGCAAPPPS 540 AAGLSGAADA AEAGQIHVMT EVYLGQQICN VVACEGPERT ERHETIGQWR GRMEAAGFEP 600 VHIGSNAFKQ ASMLLAVFAG GDGYRVEEKD GCLMLGWHTR PLITTSAWRL ADSSPRSAAA 660 PPN* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5b3g_A | 4e-61 | 251 | 657 | 26 | 382 | Protein SCARECROW |
| 5b3h_A | 5e-61 | 251 | 657 | 25 | 381 | Protein SCARECROW |
| 5b3h_D | 5e-61 | 251 | 657 | 25 | 381 | Protein SCARECROW |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcriptional regulator that acts as a repressor of the gibberellin (GA) signaling pathway. Probably acts by participating in large multiprotein complexes that repress transcription of GA-inducible genes. Upon GA application, it is degraded by the proteasome, allowing the GA signaling pathway. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00611 | ChIP-seq | Transfer from AT2G01570 | Download |
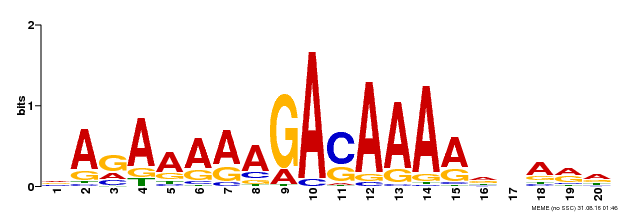
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010926472.1 | 0.0 | DELLA protein SLR1 | ||||
| Swissprot | Q8S4W7 | 0.0 | GAI1_VITVI; DELLA protein GAI1 | ||||
| TrEMBL | A0A1D1ZJ96 | 0.0 | A0A1D1ZJ96_9ARAE; DELLA protein GAI1 | ||||
| STRING | XP_008809806.1 | 0.0 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP998 | 38 | 138 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G01570.1 | 0.0 | GRAS family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Spipo15G0027800 |



