 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Spipo18G0016100 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Alismatales; Araceae; Lemnoideae; Spirodela
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 568aa MW: 62399.5 Da PI: 6.7066 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 39.7 | 8.7e-13 | 417 | 463 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCC.C...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPk.askapskKlsKaeiLekAveYIksLq 55
+h e+Er+RR+++N++f Lr ++P+ + K++Ka+ L A+ YI +Lq
Spipo18G0016100 417 NHVEAERQRREKLNQRFYALRAVVPNiS------KMDKASLLGDAIAYITDLQ 463
799***********************66......*****************99 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF14215 | 9.9E-52 | 50 | 235 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| SuperFamily | SSF55781 | 5.89E-5 | 151 | 208 | IPR029016 | GAF domain-like |
| PROSITE profile | PS50888 | 16.806 | 413 | 462 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 7.85E-19 | 416 | 479 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 3.08E-14 | 416 | 467 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 1.5E-18 | 417 | 481 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 2.9E-10 | 417 | 463 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.2E-16 | 419 | 468 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009611 | Biological Process | response to wounding | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 568 aa Download sequence Send to blast |
MGIGAGGRGF WSEEDRAMAV AVLGSQAFDF ITAGNMTAEG LTAVGSDSEL QNKLAALVDG 60 PNPTGLTWNY AIFWQISRSK SGEIILGWGD GHCREPREEE DDHLLRHRHR PQHEEEPNQA 120 MRKRVLQKLN TFFGGSDEEN FALRLDRVTD TEMFFLASMY FSFPRGEGAP GRVFASGEPL 180 WVSDCADDYC VRAFLARATG IRTVVLVPSD SGVLELGSVN SLSEDPQAMQ MITSMISHGR 240 QPKPAASSEE CPRIFGKDLN LGRTVPAVAP KLEEPPWDLQ PAGGDRLAPF PNVRKSLHVW 300 NWNQARFGSS DQTNQLSNQQ RNGVNFPQQK QQNAPAAAAA AVAAAVAAAV APPPPPPARQ 360 IDFSGVARLK SLETEKDDAE LPCKEGRPAA AAAAAGEERR PRKRGRKPAN GREEPLNHVE 420 AERQRREKLN QRFYALRAVV PNISKMDKAS LLGDAIAYIT DLQKKLKEME SEREMMDQRR 480 RAAECCPEIE VQTVGDEVVV KVSCPLETHP VSKVIHVLKE AQMNVVESKL SAGKDMVFHT 540 FVVKSHGSEQ MVKDRLVAAF SRESHPS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5gnj_A | 6e-29 | 411 | 473 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_B | 6e-29 | 411 | 473 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_E | 6e-29 | 411 | 473 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_F | 6e-29 | 411 | 473 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_G | 6e-29 | 411 | 473 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_I | 6e-29 | 411 | 473 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_M | 6e-29 | 411 | 473 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_N | 6e-29 | 411 | 473 | 2 | 64 | Transcription factor MYC2 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 398 | 406 | RRPRKRGRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that negatively regulates jasmonate (JA) signaling (PubMed:30610166). Negatively regulates JA-dependent response to wounding, JA-induced expression of defense genes, JA-dependent responses against herbivorous insects, and JA-dependent resistance against Botrytis cinerea infection (PubMed:30610166). Plays a positive role in resistance against the bacterial pathogen Pseudomonas syringae pv tomato DC3000 (PubMed:30610166). {ECO:0000269|PubMed:30610166}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00101 | PBM | Transfer from AT2G46510 | Download |
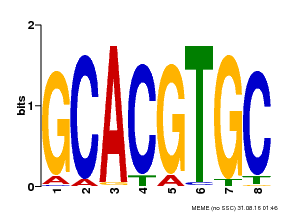
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by wounding, feeding with herbivorous insects, infection with the fungal pathogen Botrytis cinerea and infection with the bacterial pathogen Pseudomonas syringae pv tomato DC3000. {ECO:0000269|PubMed:30610166}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010242396.1 | 0.0 | PREDICTED: transcription factor bHLH13-like | ||||
| Refseq | XP_010242398.1 | 0.0 | PREDICTED: transcription factor bHLH13-like | ||||
| Refseq | XP_010242399.1 | 0.0 | PREDICTED: transcription factor bHLH13-like | ||||
| Refseq | XP_019051449.1 | 0.0 | PREDICTED: transcription factor bHLH13-like | ||||
| Swissprot | A0A3Q7ELQ2 | 0.0 | MTB1_SOLLC; Transcription factor MTB1 | ||||
| TrEMBL | A0A1D1Y608 | 0.0 | A0A1D1Y608_9ARAE; Transcription factor bHLH13 | ||||
| STRING | XP_010242395.1 | 0.0 | (Nelumbo nucifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP5174 | 38 | 60 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G01260.3 | 2e-88 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Spipo18G0016100 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



