 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Spipo18G0031000 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Alismatales; Araceae; Lemnoideae; Spirodela
|
||||||||
| Family | RAV | ||||||||
| Protein Properties | Length: 363aa MW: 39841.6 Da PI: 9.5312 | ||||||||
| Description | RAV family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 43.6 | 7.2e-14 | 60 | 108 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
s+ykGV + +grW A+I++ ++r++lg+f + eAaka++aa+++++g
Spipo18G0031000 60 SQYKGVVPQP-NGRWGAQIYEK-----HQRVWLGTFNEEAEAAKAYDAAAQRFRG 108
78****9888.8*********3.....5**********99*************98 PP
| |||||||
| 2 | B3 | 96.9 | 1.3e-30 | 189 | 286 | 1 | 93 |
EEEE-..-HHHHTT-EE--HHH.HTT.....---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSSE CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh.....ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrse 89
f+k+ltpsdv+k++rlv+pk++ae+h +g+ +++ + +ed+ g++W+++++y+++s++yvltkGW++Fvk++ Lk+gD v F+++ e
Spipo18G0031000 189 FDKALTPSDVGKLNRLVIPKQHAEKHfplqvNGPACKGVLVNFEDPCGKVWRFRYSYWNSSQSYVLTKGWSRFVKEKALKAGDLVGFHRSTGPE 282
89***************************998888899**************************************************775555 PP
E..E CS
B3 90 felv 93
++l+
Spipo18G0031000 283 RQLF 286
5555 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 3.42E-13 | 60 | 115 | No hit | No description |
| SuperFamily | SSF54171 | 9.81E-17 | 60 | 116 | IPR016177 | DNA-binding domain |
| PROSITE profile | PS51032 | 19.638 | 61 | 116 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 2.1E-20 | 61 | 116 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 4.9E-28 | 61 | 122 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 4.1E-9 | 61 | 108 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:2.40.330.10 | 3.9E-38 | 185 | 292 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 1.57E-30 | 187 | 288 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 2.17E-29 | 187 | 278 | No hit | No description |
| Pfam | PF02362 | 2.7E-27 | 189 | 286 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 5.5E-23 | 189 | 293 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 13.352 | 189 | 292 | IPR003340 | B3 DNA binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 363 aa Download sequence Send to blast |
MDSSCIEEAS SESERVAAPA PVASLSFAIP QKLQRLGSGG SVVLDPELCG EAESRKLPSS 60 QYKGVVPQPN GRWGAQIYEK HQRVWLGTFN EEAEAAKAYD AAAQRFRGRD AVTNFKPLSE 120 TDPEGAAELL FLRSHSKSEI VDMLRKHTYH DELLQSKRAI ALTSSAELGS TGCLFRSDRP 180 TAAAREKLFD KALTPSDVGK LNRLVIPKQH AEKHFPLQVN GPACKGVLVN FEDPCGKVWR 240 FRYSYWNSSQ SYVLTKGWSR FVKEKALKAG DLVGFHRSTG PERQLFIDMK PRVAAAVPGV 300 LPVKADSRPL QVMRLFGVNI VNVPGGGHRE VDESSAGKRL LEIKFLPPAQ LCKRQCVGTL 360 CV* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wid_A | 4e-52 | 186 | 292 | 11 | 117 | DNA-binding protein RAV1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways (By similarity). Transcriptional repressor of flowering time on long day plants. Acts directly on FT expression by binding 5'-CAACA-3' and 5'-CACCTG-3 sequences (Probable). Functionally redundant with TEM1. {ECO:0000250, ECO:0000269|PubMed:18718758, ECO:0000305}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00025 | PBM | Transfer from AT1G68840 | Download |
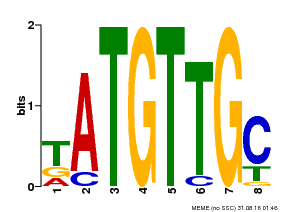
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009400502.1 | 1e-155 | PREDICTED: AP2/ERF and B3 domain-containing transcription repressor RAV2-like | ||||
| Swissprot | P82280 | 1e-131 | RAV2_ARATH; AP2/ERF and B3 domain-containing transcription repressor RAV2 | ||||
| TrEMBL | A0A3S3QJR9 | 1e-154 | A0A3S3QJR9_9MAGN; AP2/ERF and B3 domain-containing protein | ||||
| STRING | XP_008791228.1 | 1e-154 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP456 | 38 | 203 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G68840.2 | 1e-121 | related to ABI3/VP1 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Spipo18G0031000 |



