 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Spipo1G0060400 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Alismatales; Araceae; Lemnoideae; Spirodela
|
||||||||
| Family | BBR-BPC | ||||||||
| Protein Properties | Length: 336aa MW: 37407.5 Da PI: 9.9961 | ||||||||
| Description | BBR-BPC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GAGA_bind | 383.5 | 3.2e-117 | 1 | 335 | 1 | 301 |
GAGA_bind 1 mdddgsre..rnkg.yye........paaslkenlglqlmssiaerdakirernlalsekkaavaerdmaflqrdkalaernkalverdnkllal 84
mdd+g+r+ r+k +y+ p++++k++++++lm+++aerd++i+ernlalsekk a+aerdma lqrd+a+ ern+a++erd++++al
Spipo1G0060400 1 MDDSGQRDngRQKQdQYKgvhtqwmmPQHQMKDHQTIKLMTIMAERDNAIQERNLALSEKKTALAERDMAILQRDAAIGERNNAIMERDSAIAAL 95
99***99999999999*9******99778****************************************************************** PP
GAGA_bind 85 llvensla.....salpvgvqvlsgtksidslqq.lse..pqledsavelreeeklealpieeaaeeakekkkkkkrqrakkpkekkakkkkkks 171
++++++ + +p+g+++ +g+k+i+++q + +ql+d +++ r+++++ea+pi+ e+a +++++k++ r++++ ++ k++ k s
Spipo1G0060400 96 EYARENGMsgnggPGCPPGCTIPRGSKHIHHHQLsNMHppSQLSDASYGHRDMHISEAFPISVYSENAVKARRVKRS-RKEVKVQPGPKRAPK-S 188
998777666988889******************95445899*********************999999888888733.333333333333333.2 PP
GAGA_bind 172 ekskkkvkkesader..................skaekksidlvlngvslDestlPvPvCsCtGalrqCYkWGnGGWqSaCCtttiSvyPLPvst 248
+k+++ ++ ++ + +k+e+k++dl+ln+v++De+t+P+PvCsCtG+l+qCYkWGnGGWqSaCCtttiS+yPLPv++
Spipo1G0060400 189 QKKRRGGGEDLNK-HltlakghggedlnkevsaMKHEWKNQDLGLNHVAYDETTMPAPVCSCTGTLHQCYKWGNGGWQSACCTTTISMYPLPVMP 282
2222222222222.134667889999********************************************************************* PP
GAGA_bind 249 krrgaRiagrKmSqgafkklLekLaaeGydlsnpvDLkdhWAkHGtnkfvtir 301
++r+aR++grKmS++af+klL++LaaeG+dl+ p+DLkdhWAkHGtn+++ti+
Spipo1G0060400 283 NKRHARVGGRKMSGSAFTKLLSRLAAEGHDLAMPLDLKDHWAKHGTNRYITIK 335
****************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01226 | 1.3E-155 | 1 | 335 | IPR010409 | GAGA-binding transcriptional activator |
| Pfam | PF06217 | 2.0E-102 | 1 | 335 | IPR010409 | GAGA-binding transcriptional activator |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 336 aa Download sequence Send to blast |
MDDSGQRDNG RQKQDQYKGV HTQWMMPQHQ MKDHQTIKLM TIMAERDNAI QERNLALSEK 60 KTALAERDMA ILQRDAAIGE RNNAIMERDS AIAALEYARE NGMSGNGGPG CPPGCTIPRG 120 SKHIHHHQLS NMHPPSQLSD ASYGHRDMHI SEAFPISVYS ENAVKARRVK RSRKEVKVQP 180 GPKRAPKSQK KRRGGGEDLN KHLTLAKGHG GEDLNKEVSA MKHEWKNQDL GLNHVAYDET 240 TMPAPVCSCT GTLHQCYKWG NGGWQSACCT TTISMYPLPV MPNKRHARVG GRKMSGSAFT 300 KLLSRLAAEG HDLAMPLDLK DHWAKHGTNR YITIK* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional regulator that specifically binds to GA-rich elements (GAGA-repeats) present in regulatory sequences of genes involved in developmental processes. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00540 | DAP | Transfer from AT5G42520 | Download |
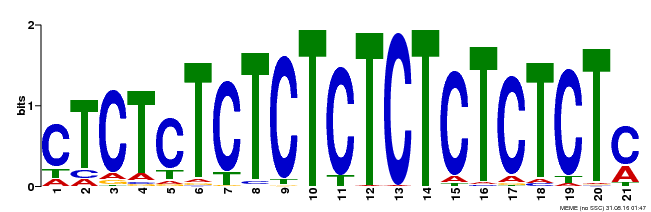
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008803316.1 | 1e-163 | barley B recombinant-like protein D isoform X2 | ||||
| Refseq | XP_010933934.1 | 1e-164 | barley B recombinant-like protein D | ||||
| Swissprot | Q5VSA8 | 1e-136 | BBRD_ORYSJ; Barley B recombinant-like protein D | ||||
| TrEMBL | A0A1D1XWN0 | 0.0 | A0A1D1XWN0_9ARAE; Barley B recombinant-like protein D | ||||
| STRING | XP_008803315.1 | 1e-162 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6226 | 37 | 56 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G42520.1 | 1e-113 | basic pentacysteine 6 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Spipo1G0060400 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



