 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Spipo1G0106700 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Alismatales; Araceae; Lemnoideae; Spirodela
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 337aa MW: 34963.9 Da PI: 7.3428 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 38.1 | 3.5e-12 | 96 | 140 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+WT++E+ ++ + k+lG+g+W+ I+r + Rt+ q+ s+ qky
Spipo1G0106700 96 PWTEDEHRMFLLGLKKLGKGDWRGISRNFVVSRTPTQVASHAQKY 140
7*******************************************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 18.058 | 89 | 145 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 2.09E-16 | 91 | 146 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.0E-18 | 92 | 144 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 1.6E-9 | 93 | 143 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.3E-10 | 94 | 139 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 0.00349 | 96 | 141 | No hit | No description |
| Pfam | PF00249 | 1.4E-9 | 96 | 140 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000122 | Biological Process | negative regulation of transcription from RNA polymerase II promoter | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0030307 | Biological Process | positive regulation of cell growth | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:2000469 | Biological Process | negative regulation of peroxidase activity | ||||
| GO:0000976 | Molecular Function | transcription regulatory region sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 337 aa Download sequence Send to blast |
MTRRCSHCSH NGHNSRTCPN RGVKLFGVRL TDGSIRKSAS MGNLSHYAGS NGGGGGSSPA 60 IDGHEVGGGA AAEGYASEDF VPGSSSGCRE RKKGIPWTED EHRMFLLGLK KLGKGDWRGI 120 SRNFVVSRTP TQVASHAQKY FIRQTNASRR KRRSSLFDII PDEVESPQLL AAGCREAENQ 180 AEDAPPEAVP IDEECESMGS TNSTEAGPGL PKPEDPLCHP VMVPAYFSPF FQLSYPVWPA 240 GYSGGGGGGA VATEPHEIVK PTAVHSKAPI NVEELVGISK LTLGDAAPSP LSLKLAGGPD 300 RQSAFHANSS AAAAAAAGAG SSDVGSSASP NPIHAL* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription repressor that binds to 5'-TATCCA-3' elements in gene promoters. Contributes to the sugar-repressed transcription of promoters containing SRS or 5'-TATCCA-3' elements. Transcription repressor involved in a cold stress response pathway that confers cold tolerance. Suppresses the DREB1-dependent signaling pathway under prolonged cold stress. DREB1 responds quickly and transiently while MYBS3 responds slowly to cold stress. They may act sequentially and complementarily for adaptation to short- and long-term cold stress (PubMed:20130099). {ECO:0000269|PubMed:12172034, ECO:0000269|PubMed:20130099}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00551 | DAP | Transfer from AT5G47390 | Download |
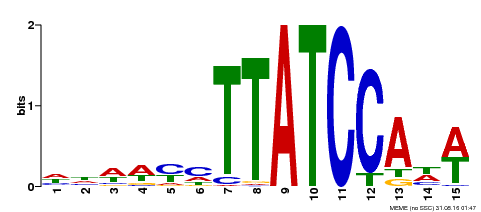
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by sucrose and gibberellic acid (GA) (PubMed:12172034). Induced by cold stress in roots and shoots. Induced by salt stress in shoots. Down-regulated by abscisic aci (ABA) in shoots (PubMed:20130099). {ECO:0000269|PubMed:12172034, ECO:0000269|PubMed:20130099}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_023552452.1 | 1e-143 | transcription factor MYBS3 | ||||
| Swissprot | Q7XC57 | 1e-123 | MYBS3_ORYSJ; Transcription factor MYBS3 | ||||
| TrEMBL | A0A2I4EIL8 | 1e-142 | A0A2I4EIL8_JUGRE; myb-like protein G | ||||
| STRING | XP_008448819.1 | 1e-140 | (Cucumis melo) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1939 | 37 | 91 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G47390.1 | 1e-100 | MYB_related family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Spipo1G0106700 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



