 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Spipo1G0117000 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Alismatales; Araceae; Lemnoideae; Spirodela
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 336aa MW: 36320.9 Da PI: 8.8065 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 53.5 | 5.7e-17 | 3 | 50 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+ Ed +l d+vk++G g+W+++ r g+ R++k+c++rw ++l
Spipo1G0117000 3 KGPWTAAEDAILTDYVKRHGEGNWNAVQRNSGLLRCGKSCRLRWANHL 50
79******************************99**********9986 PP
| |||||||
| 2 | Myb_DNA-binding | 51.5 | 2.2e-16 | 56 | 99 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
+g++++eE+ l+++++++lG++ W++ a+ ++ gRt++++k++w++
Spipo1G0117000 56 KGPFSPEEERLILELHAKLGNK-WARMAAQLP-GRTDNEIKNYWNT 99
79********************.*********.***********97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 15.932 | 1 | 50 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 4.49E-30 | 2 | 97 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 8.7E-14 | 2 | 52 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.9E-14 | 3 | 50 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 7.0E-23 | 4 | 57 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 6.09E-11 | 5 | 50 | No hit | No description |
| PROSITE pattern | PS00175 | 0 | 18 | 27 | IPR001345 | Phosphoglycerate/bisphosphoglycerate mutase, active site |
| PROSITE profile | PS51294 | 26.043 | 51 | 105 | IPR017930 | Myb domain |
| SMART | SM00717 | 4.8E-16 | 55 | 103 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.6E-15 | 56 | 99 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 9.8E-26 | 58 | 104 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 2.19E-12 | 58 | 99 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009555 | Biological Process | pollen development | ||||
| GO:0009789 | Biological Process | positive regulation of abscisic acid-activated signaling pathway | ||||
| GO:0043068 | Biological Process | positive regulation of programmed cell death | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0090406 | Cellular Component | pollen tube | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003824 | Molecular Function | catalytic activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 336 aa Download sequence Send to blast |
MKKGPWTAAE DAILTDYVKR HGEGNWNAVQ RNSGLLRCGK SCRLRWANHL RPNLRKGPFS 60 PEEERLILEL HAKLGNKWAR MAAQLPGRTD NEIKNYWNTR IKRRQRAGLP LYPLDIQRQA 120 ALHHHHRPSA GAAPLSFLDR AELGVAPTAG TSSLMVNPGH CLSFIVPSPA GETGLQQRGG 180 AFPDDGVGGF SMAFSSNTVA PPPSQNAVFL LNALGLYELS SPSPQHHRQS FSVKMEPPSS 240 QFFPEVPTRR SESDLLDSLL FDGAAVMDGG GSADKCGNVG GDHRRGHNFL PEPTAENFSY 300 VNENAGGGNS QGLVLGDCGG KWDYSKLPLS LPGTC* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 2e-30 | 3 | 104 | 7 | 107 | B-MYB |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00287 | DAP | Transfer from AT2G32460 | Download |
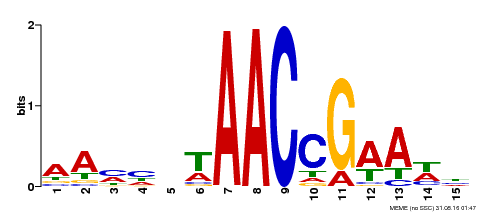
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_026392638.1 | 3e-76 | transcription factor MYB97-like | ||||
| Refseq | XP_026392639.1 | 3e-76 | transcription factor MYB97-like | ||||
| Refseq | XP_026410059.1 | 3e-76 | transcription factor MYB97-like | ||||
| TrEMBL | A0A200QBH3 | 7e-79 | A0A200QBH3_9MAGN; SANT/Myb domain | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP7420 | 37 | 50 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G32460.1 | 2e-72 | myb domain protein 101 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Spipo1G0117000 |



