 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Spipo23G0035700 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Alismatales; Araceae; Lemnoideae; Spirodela
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 214aa MW: 24255.8 Da PI: 10.2254 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 17.7 | 1e-05 | 65 | 87 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
+ C++C+k F r nL+ H r H
Spipo23G0035700 65 FICEICNKGFQRDQNLQLHRRGH 87
89*******************88 PP
| |||||||
| 2 | zf-C2H2 | 16.4 | 2.6e-05 | 142 | 164 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
+kC +C+k + +s+ k Hi+ +
Spipo23G0035700 142 WKCDKCSKKYAVQSDWKAHIKIC 164
58******************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:3.30.160.60 | 4.4E-6 | 64 | 87 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 2.62E-8 | 64 | 87 | No hit | No description |
| SMART | SM00355 | 0.022 | 65 | 87 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.845 | 65 | 87 | IPR007087 | Zinc finger, C2H2 |
| Pfam | PF12171 | 2.0E-5 | 65 | 87 | IPR022755 | Zinc finger, double-stranded RNA binding |
| PROSITE pattern | PS00028 | 0 | 67 | 87 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 78 | 107 | 137 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 5.7E-6 | 130 | 163 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 2.62E-8 | 137 | 162 | No hit | No description |
| SMART | SM00355 | 83 | 142 | 162 | IPR015880 | Zinc finger, C2H2-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010075 | Biological Process | regulation of meristem growth | ||||
| GO:0045604 | Biological Process | regulation of epidermal cell differentiation | ||||
| GO:0048364 | Biological Process | root development | ||||
| GO:0051302 | Biological Process | regulation of cell division | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 214 aa Download sequence Send to blast |
SPHLPWDSIM NQKTLTLIRS SSQRQPGRPG SSPELQSLDT SRVNTDPDAE VIALSPKTLM 60 ATNRFICEIC NKGFQRDQNL QLHRRGHNLP WKLKQRTGKE VVRKKVYVCP EKSCVHHDPS 120 RALGDLTGIK KHYSRKHGEK KWKCDKCSKK YAVQSDWKAH IKICGTREYK CDCGTLFSRK 180 DSFITHRAFC DALTEESARL SSTAVAAATG LSFK |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5b3h_C | 1e-38 | 138 | 202 | 3 | 67 | Zinc finger protein JACKDAW |
| 5b3h_F | 1e-38 | 138 | 202 | 3 | 67 | Zinc finger protein JACKDAW |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 91 | 104 | KLKQRTGKEVVRKK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that, together with BIB, regulates tissue boundaries and asymmetric cell division by a rapid up-regulation of 'SCARECROW' (SCR), thus controlling the nuclear localization of 'SHORT-ROOT' (SHR) and restricting its action (PubMed:17785527, PubMed:25829440). Binds DNA via its zinc fingers (PubMed:28211915, PubMed:24821766). Recognizes and binds to SCL3 promoter sequence 5'-AGACAA-3' to promotes its expression when in complex with RGA (PubMed:24821766). Confines CYCD6 expression to the cortex-endodermis initial/daughter (CEI/CEID) tissues (PubMed:25829440). Required for radial patterning and stem cell maintenance (PubMed:17785527). Counteracted by 'MAGPIE' (MGP) (PubMed:17785527). Binds to the SCR and MGP promoter sequences (PubMed:21935722). Controls position-dependent signals that regulate epidermal-cell-type patterning (PubMed:20356954). {ECO:0000269|PubMed:17785527, ECO:0000269|PubMed:20356954, ECO:0000269|PubMed:21935722, ECO:0000269|PubMed:24821766, ECO:0000269|PubMed:25829440, ECO:0000269|PubMed:28211915}. | |||||
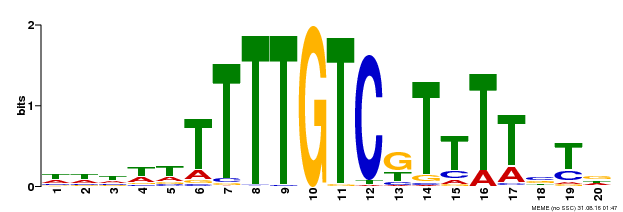
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00485 | DAP | Transfer from AT5G03150 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Not regulated by SCR and SHR. {ECO:0000269|PubMed:21935722}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006480617.1 | 1e-112 | zinc finger protein JACKDAW | ||||
| Swissprot | Q700D2 | 1e-106 | IDD10_ARATH; Zinc finger protein JACKDAW | ||||
| TrEMBL | A0A067EAZ3 | 1e-111 | A0A067EAZ3_CITSI; Uncharacterized protein | ||||
| STRING | XP_006480617.1 | 1e-112 | (Citrus sinensis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP202 | 38 | 320 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G03150.1 | 1e-97 | C2H2-like zinc finger protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Spipo23G0035700 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



