 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Spipo27G0007100 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Alismatales; Araceae; Lemnoideae; Spirodela
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 584aa MW: 60923.9 Da PI: 5.1305 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 48.2 | 1.9e-15 | 406 | 452 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
+++ +E+rRRdriN+++ L+el+P++ K++Ka++L +A+eY+k Lq
Spipo27G0007100 406 TNSVNEQRRRDRINEKMRALQELIPNC-----NKVDKASVLDEAIEYLKTLQ 452
68899*********************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50888 | 16.456 | 402 | 451 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 1.76E-14 | 405 | 456 | No hit | No description |
| Pfam | PF00010 | 1.1E-12 | 406 | 452 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 1.44E-17 | 406 | 461 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 1.6E-18 | 408 | 460 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.5E-14 | 408 | 457 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0031539 | Biological Process | positive regulation of anthocyanin metabolic process | ||||
| GO:0080113 | Biological Process | regulation of seed growth | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 584 aa Download sequence Send to blast |
MQGQSNRAAK SSSPTALPSF AGKAQERDGS RVASRYGSAD SAGKDFHAVG SLGQLGFGGG 60 QQDSDMAPWA SYPIEDYCLE FLSEFSAPNP SSLPATVPAS DRCISYGEAA LPGENGGGAR 120 GSGQFFSPPE ESLPLFPRIR SKDSEFSPHQ LPGGDLQAGK TQRHEQAPVK VAQPICGPGL 180 ANFSHFLRPS AAQAKAAAVV AEAAADLPNF DVRAGENAST AASSNPLEST LINPTSCSRS 240 FSQFHAESAP GSIAWADLDG ASAEQSGGDG EEDNSRLQNQ IDSASNPRPS AESPTCKSSS 300 FTGGAAADKH SEPVVASPSI CSASDAGAAP NDAQGKGKRK NQDAEDSVCH SEDLEDDSAE 360 AKKPSPGSKR SRAAEVHNLS ERVSPPSVAA AFSDALRNEF ISLGVTNSVN EQRRRDRINE 420 KMRALQELIP NCNKVDKASV LDEAIEYLKT LQMQLQIMSM GGGIYMPPMM VPAGIRAPHM 480 APFPPMGLGM GMGMGMGMGV GMGFGMGMVD ANGSPLVPAP PPPQYPCASV PATWNGILFC 540 LPNVIELNAI NLLALRYHSE LINSHFHICG SMQPCGESPH KVG* |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00081 | ChIP-seq | Transfer from AT1G09530 | Download |
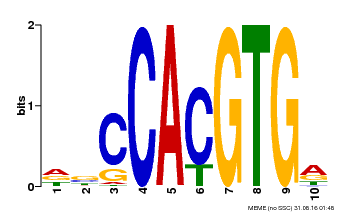
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3527 | 37 | 75 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09530.2 | 7e-39 | phytochrome interacting factor 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Spipo27G0007100 |



