 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Spipo2G0020700 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Alismatales; Araceae; Lemnoideae; Spirodela
|
||||||||
| Family | ARR-B | ||||||||
| Protein Properties | Length: 694aa MW: 73986.3 Da PI: 6.7076 | ||||||||
| Description | ARR-B family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 90.3 | 1.7e-28 | 222 | 275 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
kpr++W+ eLH++Fv+av+qL G++kA+Pk+ilelm+v+gLt+e+v+SHLQk+Rl
Spipo2G0020700 222 KPRVVWSVELHQQFVNAVNQL-GIDKAVPKRILELMNVPGLTRENVASHLQKFRL 275
79*******************.********************************8 PP
| |||||||
| 2 | Response_reg | 77.1 | 6.2e-26 | 37 | 145 | 1 | 109 |
EEEESSSHHHHHHHHHHHHHTTCEEEEEESSHHHHHHHHHHHH..ESEEEEESSCTTSEHHHHHHHHHHHTTTSEEEEEESTTTHHHHHHHHHTT CS
Response_reg 1 vlivdDeplvrellrqalekegyeevaeaddgeealellkekd..pDlillDiempgmdGlellkeireeepklpiivvtahgeeedalealkaG 93
vl+vdD+ + +++l q+l+k +y +v+++ +++al +l+e++ +Dl++ D+ mp+mdG++ll+ e++lp+i+++a e++ + +k G
Spipo2G0020700 37 VLVVDDDMTCLKILDQMLRKCEY-NVTVCPRATSALSILRERKssFDLVISDVHMPDMDGFKLLELVGL-EMDLPVIMMSADRRTEMVMKGIKHG 129
89*********************.***************988888*******************87754.558********************** PP
ESEEEESS--HHHHHH CS
Response_reg 94 akdflsKpfdpeelvk 109
a d+l Kp+ eel +
Spipo2G0020700 130 ACDYLIKPVRFEELKN 145
*************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:3.40.50.2300 | 2.0E-42 | 34 | 171 | No hit | No description |
| SuperFamily | SSF52172 | 6.26E-36 | 34 | 165 | IPR011006 | CheY-like superfamily |
| SMART | SM00448 | 2.9E-29 | 35 | 147 | IPR001789 | Signal transduction response regulator, receiver domain |
| PROSITE profile | PS50110 | 42.895 | 36 | 151 | IPR001789 | Signal transduction response regulator, receiver domain |
| Pfam | PF00072 | 1.2E-23 | 37 | 145 | IPR001789 | Signal transduction response regulator, receiver domain |
| CDD | cd00156 | 2.24E-26 | 38 | 150 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 5.2E-30 | 219 | 280 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 11.313 | 219 | 278 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.15E-19 | 219 | 279 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 5.9E-25 | 222 | 275 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 6.6E-7 | 224 | 273 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009873 | Biological Process | ethylene-activated signaling pathway | ||||
| GO:0010082 | Biological Process | regulation of root meristem growth | ||||
| GO:0010119 | Biological Process | regulation of stomatal movement | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0010380 | Biological Process | regulation of chlorophyll biosynthetic process | ||||
| GO:0031537 | Biological Process | regulation of anthocyanin metabolic process | ||||
| GO:0080022 | Biological Process | primary root development | ||||
| GO:0080036 | Biological Process | regulation of cytokinin-activated signaling pathway | ||||
| GO:0080113 | Biological Process | regulation of seed growth | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 694 aa Download sequence Send to blast |
MAAMQKLGVA NLNAGNGLGL SKTEEDVPDQ FPAGLRVLVV DDDMTCLKIL DQMLRKCEYN 60 VTVCPRATSA LSILRERKSS FDLVISDVHM PDMDGFKLLE LVGLEMDLPV IMMSADRRTE 120 MVMKGIKHGA CDYLIKPVRF EELKNIWQHV VRKKWNENKD HEQSGSLEDS DKHRRGTDEA 180 EYTSSVNDAN DCNWKSQKKK RDAKEEEDDG ELENDDPSTS KKPRVVWSVE LHQQFVNAVN 240 QLGIDKAVPK RILELMNVPG LTRENVASHL QKFRLYLKRL SGVAQHQGAL HSSMCGPVDS 300 GTKVGPLSRL DIQSLAASGQ IPPQTLAALH AELLGRPSTG MVLPGLEQPV ILQPSLQGPK 360 CIPVDHREVT FGKPLMKCPS SMSKHFQPAG ITVEDMSPSG FGSWVPSHLS PVGNASIGGL 420 NGPQNGNMLV QMLQQQQQQQ PPPQPLPSLA EASHLPMNVQ SSCLVSPSPP PAGFQIGNNP 480 FSIGQNAAVL LRSQMSKGLQ AGGGAATINQ THPGYGGPAM DHMLLPQSNA LPVGVGQVVA 540 EGDLKTAVMQ SGYFHGSPVP PAAMSAAGSS PCDGWRGSAL SMNPAGRLAG LVPSLGDAHG 600 PARATAALGG AAESQGRNLG FVGKDTGIIP SRFAADDVEP PPPPALNHNR VGGSGDTGRA 660 QEELHLGFME APKVANPGLP HFSPADVMSV LSK* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1irz_A | 2e-22 | 218 | 281 | 1 | 64 | ARR10-B |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that binds specific DNA sequence. Functions as a response regulator involved in His-to-Asp phosphorelay signal transduction system. Phosphorylation of the Asp residue in the receiver domain activates the ability of the protein to promote the transcription of target genes. May directly activate some type-A response regulators in response to cytokinins. {ECO:0000250|UniProtKB:Q940D0}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00054 | PBM | Transfer from AT4G16110 | Download |
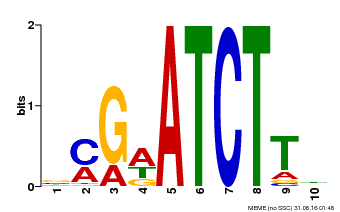
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010271366.1 | 0.0 | PREDICTED: two-component response regulator ORR21-like isoform X1 | ||||
| Refseq | XP_010271367.1 | 0.0 | PREDICTED: two-component response regulator ORR21-like isoform X1 | ||||
| Swissprot | A2XE31 | 1e-163 | ORR21_ORYSI; Two-component response regulator ORR21 | ||||
| TrEMBL | A0A1D1XD67 | 0.0 | A0A1D1XD67_9ARAE; Two-component response regulator ARR2 (Fragment) | ||||
| STRING | XP_010271366.1 | 0.0 | (Nelumbo nucifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP5796 | 37 | 54 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16110.1 | 1e-119 | response regulator 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Spipo2G0020700 |



