 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Spipo2G0052600 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Alismatales; Araceae; Lemnoideae; Spirodela
|
||||||||
| Family | M-type_MADS | ||||||||
| Protein Properties | Length: 227aa MW: 25920 Da PI: 9.9194 | ||||||||
| Description | M-type_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 49.6 | 5.1e-16 | 11 | 51 | 3 | 43 |
--SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TT CS
SRF-TF 3 ienksnrqvtfskRrngilKKAeELSvLCdaevaviifsst 43
ienk+ r tf kR++ ++KK ELS+LCd+++a++i s+
Spipo2G0052600 11 IENKTSRRQTFKKRKTCLMKKIGELSILCDVQAAMVIHSPY 51
9************************************9986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50066 | 16.848 | 1 | 49 | IPR002100 | Transcription factor, MADS-box |
| SMART | SM00432 | 3.3E-19 | 1 | 60 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 2.5E-10 | 3 | 23 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00266 | 6.59E-24 | 3 | 85 | No hit | No description |
| SuperFamily | SSF55455 | 4.45E-22 | 3 | 93 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 2.3E-16 | 11 | 52 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 2.5E-10 | 23 | 38 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 2.5E-10 | 38 | 59 | IPR002100 | Transcription factor, MADS-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0043078 | Cellular Component | polar nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 227 aa Download sequence Send to blast |
MIKKKMKLQL IENKTSRRQT FKKRKTCLMK KIGELSILCD VQAAMVIHSP YDHGIQVWPS 60 KEEASSLLRR FLDAPLYERQ KHGMDQEAFL RQQINVLNSK VEKEEKKNQE LESEYLAAWF 120 LGFRGRTLKQ LTNDELNHLQ LMVEAKLEEV QARMNAILRH QQTPGGFLTY AAVEPGPSTL 180 PHGLTSFYDA VEARIRALRG PEWQREMMAS PSLPSISAFA PSSLVP* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 2 | 24 | KKKMKLQLIENKTSRRQTFKKRK |
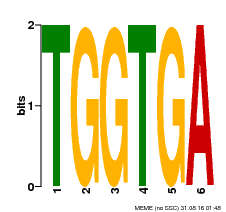
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00553 | DAP | Transfer from AT5G48670 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP363 | 36 | 202 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G26630.1 | 8e-25 | M-type_MADS family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Spipo2G0052600 |



