 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Spipo4G0054600 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Alismatales; Araceae; Lemnoideae; Spirodela
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 332aa MW: 36672.9 Da PI: 7.0643 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 135.2 | 2.1e-42 | 89 | 165 | 2 | 78 |
-SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 2 CqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
C v+gC++dls ++eyhrrhkvCevhsk+p+v+vsgleqrfCqqCsrfh+l efDe+krsCr+rL++hn+rrrk+q+
Spipo4G0054600 89 CLVDGCNSDLSGCREYHRRHKVCEVHSKTPMVMVSGLEQRFCQQCSRFHMLAEFDEGKRSCRKRLDGHNRRRRKPQP 165
**************************************************************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 4.9E-34 | 82 | 150 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 32.582 | 86 | 163 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 4.71E-40 | 87 | 168 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 1.1E-31 | 89 | 162 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0048653 | Biological Process | anther development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 332 aa Download sequence Send to blast |
MEWDLKTWDM SELEPSMRPS FAPRGSFEGQ ASGSDCSVDL KLGGSGDFGS SSASYRCRED 60 MRFSTTTVSD SSSSKRARAP SHGTQVASCL VDGCNSDLSG CREYHRRHKV CEVHSKTPMV 120 MVSGLEQRFC QQCSRFHMLA EFDEGKRSCR KRLDGHNRRR RKPQPHAMHS ANLFANFPGP 180 RFSVYSEIFP AARPESNWVA VVKTEDEVSS LHGHSSPLQF PGSYPRSYEE ERPFPFTSNR 240 TMETSVCQPL LPTITTSQGG SNDRMFHGGL AQVLDSELAL SLLSSPTRAL GINFGPMDSN 300 QISSTGFSYP GMVDEQNSPP TPPMDYVTII N* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 9e-30 | 89 | 162 | 11 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 145 | 162 | KRSCRKRLDGHNRRRRKP |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' (By similarity). May be involved in panicle development. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00555 | DAP | Transfer from AT5G50570 | Download |
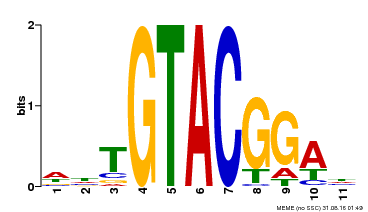
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Negatively regulated by microRNAs miR156b and miR156h. {ECO:0000305|PubMed:16861571}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010927740.1 | 1e-104 | squamosa promoter-binding-like protein 16 | ||||
| Swissprot | Q6YZE8 | 5e-62 | SPL16_ORYSJ; Squamosa promoter-binding-like protein 16 | ||||
| TrEMBL | A0A2H3YPU0 | 1e-100 | A0A2H3YPU0_PHODC; squamosa promoter-binding-like protein 16 | ||||
| STRING | XP_008801848.1 | 1e-101 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1755 | 37 | 95 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G50670.1 | 6e-48 | SBP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Spipo4G0054600 |



