 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Spipo6G0056100 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Alismatales; Araceae; Lemnoideae; Spirodela
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 196aa MW: 21368.2 Da PI: 9.3657 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 107.8 | 5.6e-34 | 44 | 98 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
kprl+Wt++LH+rF+eav+qLGG++kAtPkti++lm+++gLtl+h+kSHLQkYRl
Spipo6G0056100 44 KPRLKWTQDLHDRFIEAVNQLGGADKATPKTIMKLMGIPGLTLYHLKSHLQKYRL 98
79****************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 10.83 | 41 | 101 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.6E-31 | 42 | 99 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 1.42E-17 | 43 | 99 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 2.0E-24 | 44 | 99 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 2.6E-10 | 46 | 97 | IPR001005 | SANT/Myb domain |
| Pfam | PF14379 | 6.6E-9 | 140 | 160 | IPR025756 | MYB-CC type transcription factor, LHEQLE-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 196 aa Download sequence Send to blast |
MYGHHEGKHG ILTSRAAFHQ DRNLLLQGAN APGDAGLVLS TDAKPRLKWT QDLHDRFIEA 60 VNQLGGADKA TPKTIMKLMG IPGLTLYHLK SHLQKYRLSK NLHTQANNGT SKNASAGRQA 120 EVSGALTGTA VPSPTNKTLQ IGEALQMQIE VQKRLHEQLE NSMDLADHPK YASKEVVSVE 180 DTDWIFSFIG IKPEG* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 4e-21 | 44 | 100 | 2 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 4e-21 | 44 | 100 | 2 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_A | 4e-21 | 44 | 100 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 4e-21 | 44 | 100 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 4e-21 | 44 | 100 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 4e-21 | 44 | 100 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 4e-21 | 44 | 100 | 2 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 4e-21 | 44 | 100 | 2 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 4e-21 | 44 | 100 | 2 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 4e-21 | 44 | 100 | 2 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 4e-21 | 44 | 100 | 2 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 4e-21 | 44 | 100 | 2 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that may activate the transcription of specific genes involved in nitrogen uptake or assimilation (PubMed:15592750). Acts redundantly with MYR1 as a repressor of flowering and organ elongation under decreased light intensity (PubMed:21255164). Represses gibberellic acid (GA)-dependent responses and affects levels of bioactive GA (PubMed:21255164). {ECO:0000269|PubMed:21255164, ECO:0000305|PubMed:15592750}. | |||||
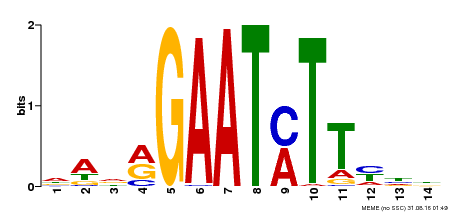
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00331 | DAP | Transfer from AT3G04030 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by nitrogen deficiency. {ECO:0000269|PubMed:15592750}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_026665294.1 | 4e-76 | myb-related protein 2-like isoform X3 | ||||
| Swissprot | Q9SQQ9 | 1e-59 | PHL9_ARATH; Myb-related protein 2 | ||||
| TrEMBL | A0A1D1Z681 | 4e-82 | A0A1D1Z681_9ARAE; Myb family transcription factor APL | ||||
| STRING | XP_008807359.1 | 6e-74 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3574 | 38 | 79 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G04030.1 | 4e-62 | G2-like family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Spipo6G0056100 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



