 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Spipo6G0070000 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Alismatales; Araceae; Lemnoideae; Spirodela
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 270aa MW: 30900.8 Da PI: 5.811 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 50.6 | 4.4e-16 | 8 | 55 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT++Ed +l+ v++lG ++W+ +a+ g++Rt+k+c++rw +yl
Spipo6G0070000 8 KGPWTEQEDRQLASSVELLGDRRWDFVAKVSGLNRTGKSCRLRWVNYL 55
79********************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 57.6 | 2.9e-18 | 61 | 106 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rgr+T++E+ l++++++++G++ W++Iar+++ gRt++++k++w++++
Spipo6G0070000 61 RGRMTPQEERLIIELHARWGNR-WSRIARRLP-GRTDNEIKNYWRTHM 106
89********************.*********.************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 16.926 | 3 | 55 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.83E-28 | 7 | 102 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.9E-14 | 7 | 57 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.6E-15 | 8 | 55 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 3.5E-20 | 9 | 62 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.08E-9 | 10 | 55 | No hit | No description |
| PROSITE profile | PS51294 | 26.124 | 56 | 110 | IPR017930 | Myb domain |
| SMART | SM00717 | 3.1E-17 | 60 | 108 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.0E-16 | 61 | 105 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 7.6E-24 | 63 | 109 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.31E-11 | 63 | 106 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 270 aa Download sequence Send to blast |
MVKEETRKGP WTEQEDRQLA SSVELLGDRR WDFVAKVSGL NRTGKSCRLR WVNYLDPCLK 60 RGRMTPQEER LIIELHARWG NRWSRIARRL PGRTDNEIKN YWRTHMRKKA QEGRTKAWSP 120 PQSSASSPSS LYLQSAPPTE APAVNPEPCG AGMEAWPREK EDDGEQEGCP MEQIWDEISG 180 LARFEEFSED EACNISCLPM SSPLWDYARD TPSWKIDEEF KMFLPFSAPV WSMEEDPANT 240 QTLPVHLHSS NAASSTCGRC GIIRELFPR* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gv2_A | 2e-27 | 5 | 110 | 1 | 105 | MYB PROTO-ONCOGENE PROTEIN |
| 1h88_C | 1e-26 | 5 | 110 | 55 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1h89_C | 1e-26 | 5 | 110 | 55 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1h8a_C | 4e-27 | 5 | 110 | 24 | 128 | MYB TRANSFORMING PROTEIN |
| 1mse_C | 3e-27 | 8 | 110 | 4 | 105 | C-Myb DNA-Binding Domain |
| 1msf_C | 3e-27 | 8 | 110 | 4 | 105 | C-Myb DNA-Binding Domain |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. {ECO:0000305}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00600 | PBM | Transfer from AT5G59780 | Download |
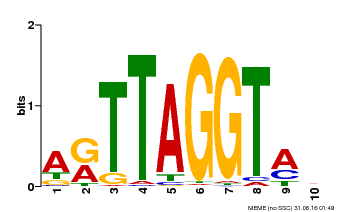
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008795619.1 | 6e-82 | myb-related protein MYBAS1-like | ||||
| Swissprot | Q53NK6 | 1e-72 | MYBA1_ORYSJ; Myb-related protein MYBAS1 | ||||
| TrEMBL | A0A2H3YA75 | 1e-80 | A0A2H3YA75_PHODC; myb-related protein MYBAS1-like | ||||
| STRING | XP_008795619.1 | 2e-81 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP636 | 37 | 169 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G59780.3 | 2e-66 | myb domain protein 59 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Spipo6G0070000 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



