 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | TRAES3BF001900070CFD_t1 | ||||||||
| Common Name | TRAES_3BF001900070CFD_c1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Triticinae; Triticum
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 343aa MW: 37353.1 Da PI: 9.8837 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 48.8 | 1.6e-15 | 273 | 318 | 5 | 51 |
CHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkelee 51
+r++r++kNRe+A rsR+RK+a++ eLe+kv Le+eN++Lkk+ +e
TRAES3BF001900070CFD_t1 273 RRQKRMIKNRESAARSRARKQAYTNELENKVSRLEEENERLKKQ-KE 318
69****************************************65.34 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 1.7E-14 | 269 | 341 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 11.702 | 271 | 316 | IPR004827 | Basic-leucine zipper domain |
| Gene3D | G3DSA:1.20.5.170 | 4.7E-15 | 271 | 316 | No hit | No description |
| CDD | cd14707 | 8.63E-25 | 273 | 322 | No hit | No description |
| Pfam | PF00170 | 6.8E-13 | 273 | 317 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 1.81E-11 | 273 | 319 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 276 | 291 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 343 aa Download sequence Send to blast |
MGSRKMASQQ GRGDAGTSQP GRGDAGTSQR GQVQSLARQG SLYSLTLDEV QNHLGEPLQS 60 MNLDELLRTV FPDDLEPDGA TTSQCVPSSS LLRQGSITMP TELSKKTVDE VWKGIQETPK 120 RSVQGSGRRK RERQPTLGEM TLEDFLVQAG VVSQGFLKDT SDVGNLGLMG RGATAAGATD 180 LTSGAQWLGQ YQQQIAASAI NTHQHVQQIV PAAYMPIQLV PQPLNVVGPG ATLGSAYSDG 240 QSTSPMISPI SDSQTPGRKR GVSGDVPNKF VERRQKRMIK NRESAARSRA RKQAYTNELE 300 NKVSRLEEEN ERLKKQKELN MMLCSVPLPE PKYQLRRTCS AAF |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 116 | 121 | ETPKRS |
| 2 | 126 | 133 | GRRKRERQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds to the embryo specification element and the ABA-responsive element (ABRE) of the Dc3 gene promoter. Could participate in abscisic acid-regulated gene expression during seed development. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00409 | DAP | Transfer from AT3G56850 | Download |
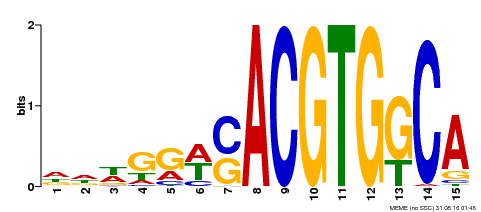
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HG670306 | 0.0 | HG670306.1 Triticum aestivum chromosome 3B, genomic scaffold, cultivar Chinese Spring. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020190745.1 | 0.0 | ABSCISIC ACID-INSENSITIVE 5-like protein 2 isoform X2 | ||||
| Swissprot | Q9LES3 | 3e-83 | AI5L2_ARATH; ABSCISIC ACID-INSENSITIVE 5-like protein 2 | ||||
| TrEMBL | A0A077S119 | 0.0 | A0A077S119_WHEAT; Uncharacterized protein | ||||
| TrEMBL | A0A446Q2V3 | 0.0 | A0A446Q2V3_TRITD; Uncharacterized protein | ||||
| STRING | Traes_3B_5D3F7382A.1 | 0.0 | (Triticum aestivum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2707 | 38 | 83 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G56850.1 | 1e-85 | ABA-responsive element binding protein 3 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



