 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | TRAES3BF002700090CFD_t1 | ||||||||
| Common Name | TRAES_3BF002700090CFD_c1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Triticinae; Triticum
|
||||||||
| Family | TCP | ||||||||
| Protein Properties | Length: 280aa MW: 30354.8 Da PI: 7.9831 | ||||||||
| Description | TCP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | TCP | 96 | 7.3e-30 | 53 | 122 | 3 | 72 |
TCP 3 gkkdrhskihTkvggRdRRvRlsaecaarfFdLqdeLGfdkdsktieWLlqqakpaikeltgtssssase 72
g+kdrhsk+ T +g+RdRRvRls+++a++++dLqd+LG+ ++sk ++WLl++a ++i++l+ +++++++
TRAES3BF002700090CFD_t1 53 GGKDRHSKVKTVKGLRDRRVRLSVQTAIQLYDLQDRLGLNQPSKVVDWLLNAARHEIDKLPPLQFPPHQV 122
78*************************************************************9999922 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03634 | 8.0E-28 | 54 | 117 | IPR005333 | Transcription factor, TCP |
| PROSITE profile | PS51369 | 29.665 | 54 | 112 | IPR017887 | Transcription factor TCP subgroup |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0030154 | Biological Process | cell differentiation | ||||
| GO:0045962 | Biological Process | positive regulation of development, heterochronic | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 280 aa Download sequence Send to blast |
MMSGNNQHQQ SAAAAAAEEE QELARKHAAA VATSRQWSSQ TESRIVRVSR VFGGKDRHSK 60 VKTVKGLRDR RVRLSVQTAI QLYDLQDRLG LNQPSKVVDW LLNAARHEID KLPPLQFPPH 120 QVDLMDHMMT SSSMPLMPHV DDKFCHFAST LARDGGVKAG ADADGGGAHH NIGRFGYHRF 180 MGLNNNSLGL VNSGMPYNFT GESWNNSSVD QSSGAGSPQV SIAAAAAAHH SAFPSLLSLA 240 PRSHQLVYYS SEADQFPVDD LGSQSLSLSS ARAFHDQTGS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5zkt_A | 4e-20 | 59 | 113 | 1 | 55 | Putative transcription factor PCF6 |
| 5zkt_B | 4e-20 | 59 | 113 | 1 | 55 | Putative transcription factor PCF6 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00065 | PBM | Transfer from AT5G60970 | Download |
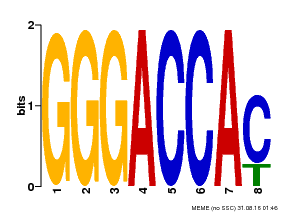
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HG670306 | 0.0 | HG670306.1 Triticum aestivum chromosome 3B, genomic scaffold, cultivar Chinese Spring. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020168249.1 | 1e-170 | uncharacterized protein LOC109753738 | ||||
| TrEMBL | A0A077RVE9 | 0.0 | A0A077RVE9_WHEAT; Uncharacterized protein | ||||
| TrEMBL | A0A446Q0A9 | 0.0 | A0A446Q0A9_TRITD; Uncharacterized protein | ||||
| STRING | MLOC_14785.2 | 1e-157 | (Hordeum vulgare) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2490 | 35 | 84 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G60970.1 | 2e-39 | TEOSINTE BRANCHED 1, cycloidea and PCF transcription factor 5 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



