 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | TRAES3BF073700150CFD_t1 | ||||||||
| Common Name | TRAES_3BF073700150CFD_c1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Triticinae; Triticum
|
||||||||
| Family | M-type_MADS | ||||||||
| Protein Properties | Length: 251aa MW: 27963.3 Da PI: 8.2103 | ||||||||
| Description | M-type_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 54.7 | 1.2e-17 | 11 | 51 | 3 | 43 |
--SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TT CS
SRF-TF 3 ienksnrqvtfskRrngilKKAeELSvLCdaevaviifsst 43
i n s r tf kRr g++KKA+EL +LCd++++v+++++
TRAES3BF073700150CFD_t1 11 IPNVSTRRGTFKKRRRGLMKKASELAILCDVRACVLVYGEG 51
779999********************************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00432 | 8.0E-20 | 1 | 60 | IPR002100 | Transcription factor, MADS-box |
| PROSITE profile | PS50066 | 18.502 | 1 | 50 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00266 | 2.30E-29 | 2 | 85 | No hit | No description |
| SuperFamily | SSF55455 | 2.62E-23 | 3 | 95 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 8.3E-11 | 3 | 23 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 8.5E-16 | 11 | 51 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 8.3E-11 | 23 | 38 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 8.3E-11 | 38 | 59 | IPR002100 | Transcription factor, MADS-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0043078 | Cellular Component | polar nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 251 aa Download sequence Send to blast |
MARKKVTLQY IPNVSTRRGT FKKRRRGLMK KASELAILCD VRACVLVYGE GETTPEVFPS 60 HDGAVKILNI FKKMPELEQC KKMMNQEGFL RQRIDKLRDQ VHKFGREFRE QEIRTLLLKA 120 MCGNLPDLVS LNIEELTSVG WKVEMLLNGI GDRITKLQGQ PPAPHMIDSM DMGSPAINQA 180 PPAQEGWLDM VRSGGDLSAL VCSGYFGSHE ATSTSSSVGY SGGDMMQPFD LGFGWQWDTD 240 LGASSSLFLP M |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 2 | 24 | RKKVTLQYIPNVSTRRGTFKKRR |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00553 | DAP | Transfer from AT5G48670 | Download |
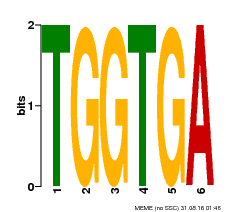
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HG670306 | 0.0 | HG670306.1 Triticum aestivum chromosome 3B, genomic scaffold, cultivar Chinese Spring. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020199163.1 | 1e-168 | agamous-like MADS-box protein AGL80 | ||||
| TrEMBL | A0A077RXW0 | 0.0 | A0A077RXW0_WHEAT; Uncharacterized protein | ||||
| STRING | EMT02802 | 1e-134 | (Aegilops tauschii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP363 | 36 | 202 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G48670.1 | 9e-31 | AGAMOUS-like 80 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



