 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | TRAES3BF094200030CFD_t1 | ||||||||
| Common Name | TRAES_3BF094200030CFD_c1, TRAES_3BF107400040CFD_c1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Triticinae; Triticum
|
||||||||
| Family | ARR-B | ||||||||
| Protein Properties | Length: 584aa MW: 64858.3 Da PI: 5.1932 | ||||||||
| Description | ARR-B family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 81.7 | 8.5e-26 | 191 | 244 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
k+r++W+ +LH++Fv+av+q+ G +k Pk+il+lm+v+gLt+e+v+SHLQkYRl
TRAES3BF094200030CFD_t1 191 KARVVWSVDLHQKFVNAVNQI-GFDKVGPKKILDLMNVPGLTRENVASHLQKYRL 244
68*******************.9*******************************8 PP
| |||||||
| 2 | Response_reg | 81.2 | 3.4e-27 | 13 | 121 | 1 | 109 |
EEEESSSHHHHHHHHHHHHHTTCEEEEEESSHHHHHHHHHHHH..ESEEEEESSCTTSEHHHHHHHHHHHTTTSEEEEEESTTTHH CS
Response_reg 1 vlivdDeplvrellrqalekegyeevaeaddgeealellkekd..pDlillDiempgmdGlellkeireeepklpiivvtahgeee 84
vl+vdD+p+ +++l+++l+k y ev+++ ++ ale+l+e++ +D+++ D++mp+mdG++ll++i e +lp+i+++ ge +
TRAES3BF094200030CFD_t1 13 VLVVDDDPTWLKILEKMLRKCSY-EVTTCGLARVALEILRERKnrFDIVISDVNMPDMDGFKLLEHIGLEM-DLPVIMMSIDGETS 96
89*********************.***************877778**********************6654.8************* PP
HHHHHHHTTESEEEESS--HHHHHH CS
Response_reg 85 dalealkaGakdflsKpfdpeelvk 109
+ + ++ Ga d+l Kp+ ++el++
TRAES3BF094200030CFD_t1 97 RVMKGVQHGACDYLLKPVRMKELRN 121
**********************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PIRSF | PIRSF036392 | 1.1E-164 | 1 | 575 | IPR017053 | Response regulator B-type, plant |
| SuperFamily | SSF52172 | 4.7E-35 | 10 | 133 | IPR011006 | CheY-like superfamily |
| Gene3D | G3DSA:3.40.50.2300 | 5.9E-44 | 10 | 144 | No hit | No description |
| SMART | SM00448 | 1.5E-31 | 11 | 123 | IPR001789 | Signal transduction response regulator, receiver domain |
| PROSITE profile | PS50110 | 42.762 | 12 | 127 | IPR001789 | Signal transduction response regulator, receiver domain |
| Pfam | PF00072 | 2.1E-24 | 13 | 122 | IPR001789 | Signal transduction response regulator, receiver domain |
| CDD | cd00156 | 1.02E-29 | 14 | 126 | No hit | No description |
| SuperFamily | SSF46689 | 1.08E-17 | 188 | 247 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 5.9E-28 | 189 | 248 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.0E-22 | 191 | 244 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 9.2E-8 | 193 | 243 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000160 | Biological Process | phosphorelay signal transduction system | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009735 | Biological Process | response to cytokinin | ||||
| GO:0010082 | Biological Process | regulation of root meristem growth | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 584 aa Download sequence Send to blast |
MADASAFPYG LRVLVVDDDP TWLKILEKML RKCSYEVTTC GLARVALEIL RERKNRFDIV 60 ISDVNMPDMD GFKLLEHIGL EMDLPVIMMS IDGETSRVMK GVQHGACDYL LKPVRMKELR 120 NIWQHVYRKK MHEVKEIEGH DSCDDLQILR YGFEGFDEKG LFMTVDSDAT RKRKDVDHGD 180 QDSSDGATAK KARVVWSVDL HQKFVNAVNQ IGFDKVGPKK ILDLMNVPGL TRENVASHLQ 240 KYRLYLGRLQ KQNEERLLGS ARQDFSTKGP SSENLNLRSS FQEQPSNTSS GYPHASQKIQ 300 GQSSVSDSQL EETKRMVPLP APDRSMNSVS SAAEPQNVAG VSPIGGVLSF KGLPVNQDRK 360 PSETMILECQ AWTGGVPAKQ FMQYPKHNHA RCDLLGDYAC LPKPDLEHPT APGHLFTPPP 420 LISMSCSTEM DARNFSDVKP ALVDCIKSFS PALTCTADSV SVQISDSVVT STDAADRKFS 480 SVEGLPTTKD CYFGQTSNQG SWLRSQEEPS IICGADFASL PEDLPGYPLQ GGVSFENVGL 540 SSIDLFHYND AMILSGLQSN WYDDQDHFSS ETTDYPLMDG CLFA |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1irz_A | 8e-18 | 190 | 249 | 4 | 63 | ARR10-B |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that binds specific DNA sequence. Functions as a response regulator involved in His-to-Asp phosphorelay signal transduction system. Phosphorylation of the Asp residue in the receiver domain activates the ability of the protein to promote the transcription of target genes. May directly activate some type-A response regulators in response to cytokinins. {ECO:0000250|UniProtKB:Q940D0}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00010 | PBM | Transfer from AT1G67710 | Download |
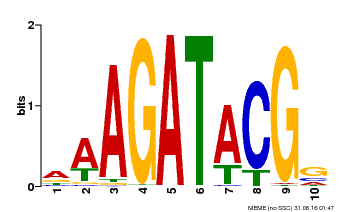
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HG670306 | 0.0 | HG670306.1 Triticum aestivum chromosome 3B, genomic scaffold, cultivar Chinese Spring. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020147288.1 | 0.0 | two-component response regulator ORR26-like | ||||
| Swissprot | Q5N6V8 | 0.0 | ORR26_ORYSJ; Two-component response regulator ORR26 | ||||
| TrEMBL | W5D4U0 | 0.0 | W5D4U0_WHEAT; Two-component response regulator | ||||
| STRING | Traes_3B_D9F27151E.2 | 0.0 | (Triticum aestivum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP5900 | 37 | 52 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G67710.1 | 1e-113 | response regulator 11 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



