 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | TRAES3BF098300010CFD_t1 | ||||||||
| Common Name | TRAES_3BF098300010CFD_c1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Triticinae; Triticum
|
||||||||
| Family | RAV | ||||||||
| Protein Properties | Length: 350aa MW: 38321.4 Da PI: 10.2104 | ||||||||
| Description | RAV family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 34.7 | 4.4e-11 | 55 | 103 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkkleg 55
s++kGV + +grW A+I++ r +r++lg+f + Aa+a++ a ++++g
TRAES3BF098300010CFD_t1 55 SRFKGVVPQP-NGRWGAQIYE------RhARVWLGTFPDQDSAARAYDVASLRYRG 103
689***9788.8*********......44*************************98 PP
| |||||||
| 2 | B3 | 92.7 | 2.6e-29 | 169 | 262 | 1 | 85 |
EEEE-..-HHHHTT-EE--HHH.HTT.........---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh.........ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkeg 77
f+k+ tpsdv+k++rlv+pk++ae+h + +++ l +ed +g++W+++++y+++s++yvltkGW++Fv+++gL +g
TRAES3BF098300010CFD_t1 169 FEKAVTPSDVGKLNRLVVPKQHAEKHfplkrtpetPTTTGKGVLLNFEDGEGKVWRFRYSYWNSSQSYVLTKGWSRFVREKGLGAG 254
89*************************9988776644445999******************************************* PP
-EEEEEE- CS
B3 78 DfvvFkld 85
D+++F+ +
TRAES3BF098300010CFD_t1 255 DSILFSCS 262
*****943 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF54171 | 3.53E-15 | 55 | 112 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 8.0E-7 | 55 | 103 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 1.89E-20 | 55 | 111 | No hit | No description |
| Gene3D | G3DSA:3.30.730.10 | 5.9E-18 | 56 | 111 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 3.6E-19 | 56 | 117 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 20.192 | 56 | 111 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:2.40.330.10 | 2.5E-37 | 164 | 281 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 4.58E-29 | 166 | 275 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 2.15E-28 | 167 | 259 | No hit | No description |
| Pfam | PF02362 | 3.9E-27 | 169 | 264 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 13.676 | 169 | 278 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 4.8E-22 | 169 | 277 | IPR003340 | B3 DNA binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 350 aa Download sequence Send to blast |
MGVEILSSMV EHSFQYSSGV STATTESGTA GTPPRPLSLP VAIADESVTS RSASSRFKGV 60 VPQPNGRWGA QIYERHARVW LGTFPDQDSA ARAYDVASLR YRGRDVAFNF PCAAVEGELA 120 FLAAHSKAEI VDMLRKQTYA DELRQGLRRG RGMGARAQPT PSWAREPLFE KAVTPSDVGK 180 LNRLVVPKQH AEKHFPLKRT PETPTTTGKG VLLNFEDGEG KVWRFRYSYW NSSQSYVLTK 240 GWSRFVREKG LGAGDSILFS CSLYEQEKQF FIDCKKNTSM NGGKSASPLP VGVTTKGEQV 300 RVVRLFGVDI SGVKRGRAAT ATAEQGLQEL FKRQCVAPGQ HSPALGAFAL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wid_A | 1e-45 | 166 | 275 | 11 | 115 | DNA-binding protein RAV1 |
| Search in ModeBase | ||||||
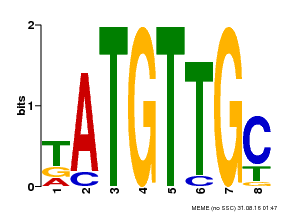
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00025 | PBM | Transfer from AT1G68840 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HG670306 | 0.0 | HG670306.1 Triticum aestivum chromosome 3B, genomic scaffold, cultivar Chinese Spring. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020191173.1 | 0.0 | AP2/ERF and B3 domain-containing protein Os01g0141000-like isoform X1 | ||||
| Refseq | XP_020200371.1 | 0.0 | AP2/ERF and B3 domain-containing protein Os01g0141000-like isoform X1 | ||||
| Refseq | XP_020201146.1 | 0.0 | AP2/ERF and B3 domain-containing protein Os01g0141000-like isoform X1 | ||||
| Swissprot | Q9AWS0 | 1e-134 | Y1410_ORYSJ; AP2/ERF and B3 domain-containing protein Os01g0141000 | ||||
| TrEMBL | W5CWS4 | 0.0 | W5CWS4_WHEAT; Uncharacterized protein | ||||
| STRING | Traes_3B_68BFA25F1.1 | 0.0 | (Triticum aestivum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP456 | 38 | 203 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G68840.2 | 4e-99 | related to ABI3/VP1 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



