 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | TRAES3BF104600030CFD_t1 | ||||||||
| Common Name | TRAES_3BF104600030CFD_c1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Triticinae; Triticum
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 604aa MW: 65992 Da PI: 8.2351 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 50.4 | 5e-16 | 66 | 112 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g WT +Ede l +av+ + g++Wk+Ia+ ++ Rt+ qc +rwqk+l
TRAES3BF104600030CFD_t1 66 KGGWTLQEDETLRKAVETFNGRSWKKIAEFFP-DRTEVQCLHRWQKVL 112
688*****************************.************986 PP
| |||||||
| 2 | Myb_DNA-binding | 63.6 | 3.9e-20 | 118 | 164 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+eEd++++d+vk++G+ W+ Iar ++ gR +kqc++rw+++l
TRAES3BF104600030CFD_t1 118 KGPWTQEEDDKIIDLVKKYGPTKWSVIARSLP-GRIGKQCRERWHNHL 164
79******************************.*************97 PP
| |||||||
| 3 | Myb_DNA-binding | 60.5 | 3.7e-19 | 170 | 213 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
+++WT+eE++ l++a++++G++ W+ Ia+ ++ gRt++++k++w++
TRAES3BF104600030CFD_t1 170 KDAWTAEEEQALINAHREYGNK-WAEIAKVLP-GRTDNSIKNHWNS 213
679*******************.*********.***********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 13.684 | 61 | 112 | IPR017930 | Myb domain |
| SMART | SM00717 | 8.1E-14 | 65 | 114 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.4E-14 | 66 | 112 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 3.23E-15 | 67 | 122 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 5.8E-21 | 68 | 124 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 2.06E-12 | 69 | 112 | No hit | No description |
| PROSITE profile | PS51294 | 31.783 | 113 | 168 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.58E-32 | 115 | 211 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 8.5E-19 | 117 | 166 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.8E-18 | 118 | 164 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 9.42E-17 | 120 | 164 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 2.0E-27 | 125 | 171 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.3E-18 | 169 | 217 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS51294 | 24.45 | 169 | 219 | IPR017930 | Myb domain |
| Pfam | PF00249 | 1.8E-17 | 170 | 213 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 3.30E-15 | 172 | 215 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 7.9E-25 | 172 | 219 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 604 aa Download sequence Send to blast |
MGAMAEAEHE GCVENRQPLA ASSESVSEGS SYGGAGGPAR MSPPVSGSVN SISGLRRTSG 60 PIRRAKGGWT LQEDETLRKA VETFNGRSWK KIAEFFPDRT EVQCLHRWQK VLNPELIKGP 120 WTQEEDDKII DLVKKYGPTK WSVIARSLPG RIGKQCRERW HNHLNPDIRK DAWTAEEEQA 180 LINAHREYGN KWAEIAKVLP GRTDNSIKNH WNSSLRKKLD VYGTRNILAI PRLVGHDDFK 240 DKQKPVASEG HLDLNTVPGI TSKNLPEIAH HSNFSSHLQS YKLDHAKAGS GFLSISFLPT 300 VQPLTSSEVS SVVNGSAVTL AAQGLESDSV RDKALEIVSV HEKGLKVDST LDTLREPGST 360 QLEAVPAKGE ESSLKNDAQS SLGPLCYQIP NMNDVAPASS SLFSEHQTVH QTSEHCRNGA 420 LSPNGCTTPT KGTMSVQLSV DSILKIAADS FPVTPSILRR RKRERPTPAS DLKFGEPNTD 480 SFYTPAGKRP TTDTPESFRT ASFLSLGSLD GLSSSARGFD VSPPYRIRAK RMCLTKAVEK 540 QLDFSSNGLG TCGSEVLNSP HQNSQSTHSL GEASIMKEKE LNGHAIQLET QTKNIAYTTN 600 MDVT |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h88_C | 4e-73 | 65 | 219 | 5 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1h89_C | 4e-73 | 65 | 219 | 5 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in abiotic stress responses (PubMed:17293435, PubMed:19279197). May play a regulatory role in tolerance to salt, cold, and drought stresses (PubMed:17293435). Transcriptional activator that binds specifically to a mitosis-specific activator cis-element 5'-(T/C)C(T/C)AACGG(T/C)(T/C)A-3', found in promoters of cyclin genes such as CYCB1-1 and KNOLLE (AC Q84R43). Positively regulates a subset of G2/M phase-specific genes, including CYCB1-1, CYCB2-1, CYCB2-2, and CDC20.1 in response to cold treatment (PubMed:19279197). {ECO:0000269|PubMed:17293435, ECO:0000269|PubMed:19279197}. | |||||
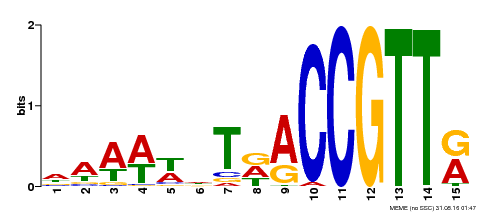
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00482 | DAP | Transfer from AT5G02320 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by cold, drought and salt stresses. {ECO:0000269|PubMed:17293435}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HQ236494 | 0.0 | HQ236494.1 Triticum aestivum MYB3R transcription factor (MYB3R1) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020192352.1 | 0.0 | myb-related protein B-like | ||||
| Swissprot | Q0JHU7 | 0.0 | MB3R2_ORYSJ; Transcription factor MYB3R-2 | ||||
| TrEMBL | A0A077S4V4 | 0.0 | A0A077S4V4_WHEAT; Uncharacterized protein | ||||
| TrEMBL | A0A446Q4Z5 | 0.0 | A0A446Q4Z5_TRITD; Uncharacterized protein | ||||
| STRING | Traes_3B_B594FC28C.2 | 0.0 | (Triticum aestivum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3099 | 38 | 80 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G02320.2 | 1e-108 | myb domain protein 3r-5 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



